
 FasterDB identifier: 13803 - VOPP1 (DKFZp564K0822, ECop, FLJ20532, GASP)
vesicular, overexpressed in cancer, prosurvival protein 1
FasterDB identifier: 13803 - VOPP1 (DKFZp564K0822, ECop, FLJ20532, GASP)
vesicular, overexpressed in cancer, prosurvival protein 1

| Gene |  |
||
|---|---|---|---|
| Symbol | VOPP1 | ||
| Synonyms | DKFZp564K0822, ECop, FLJ20532, GASP | ||
| Description | vesicular, overexpressed in cancer, prosurvival protein 1 | ||
| EnsEMBL link | ENSG00000154978 | ||
| Chromosome | 7 - reverse strand | ||
| Chromosomal location (UCSC link) |
55538307 - 55640200 (101894bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_030796 AK225755 XM_001723367 AK075391 AK314338 BC016650 AB097004 AL136610 XM_001129280 CR533483 AK090825 AK309288 AK315882 AK092992 AF395824 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 55640200 | NM_030796 |
| P2 | 1 | 95 | 55640106 | AK314338 |
| P3 | 2 | 640 | 55639561 | AK090825 |
| P4 | 3 | 19735 | 55620466 | AK309288 |
| P5 | 4 | 33848 | 55606353 | AK315882 |
| P6 | 5 | 34814 | 55605387 | AK092992 |
| P7 | 6 | 51364 | 55588837 | AF395824 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_030796 | sequence |
| AK225755 | sequence |
| XM_001723367 | sequence |
| AK075391 | sequence |
| AK314338 | sequence |
| BC016650 | sequence |
| AB097004 | sequence |
| AL136610 | sequence |
| XM_001129280 | sequence |
| CR533483 | sequence |
| AK090825 | sequence |
| AK309288 | sequence |
| AK315882 | sequence |
| AK092992 | sequence |
| AF395824 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 9 | 99653 | 55540548 | no | AK314338 | Full_length | |
| PA2 | 9 | 100305 | 55539896 | no | AK309288 | Full_length | |
| PA3 | 9 | 100540 | 55539661 | no | AK315882 | Full_length | |
| PA4 | 9 | 101894 | 55538307 | AATAAA | yes | NM_030796 | Cdna |
| PA5 | 9 | 101894 | 55538307 | AATAAA | no | AK075391 | Cdna |
| PA6 | 9 | 101894 | 55538307 | AATAAA | yes | BC016650 | Cdna |
| PA7 | 9 | 101894 | 55538307 | AATAAA | no | AB097004 | Cdna |
| PA8 | 9 | 101894 | 55538307 | AATAAA | yes | AL136610 | Cdna |
| PA9 | 9 | 101894 | 55538307 | AATAAA | no | AF395824 | Cdna |
| PA10 | 9 | 101894 | 55538307 | AATAAA | no | AK090825 | Full_length |
| PA11 | 9 | 101894 | 55538307 | AATAAA | no | AK092992 | Full_length |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 2 |
| 2 | 1 |
| 3 | 1 |
| 4 | 1 |
| 5 | 1 |
| 6 | 1 |
| Transcription termination & last exon(s) | |
| 9 | 11 |
| Exon skipping | |
| 2 | 10 |
| 3 | 11 |
| 4 | 12 |
| 5 | 13 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 237 | 55639964-55640200 | 237 | sequence | download |
| 1 | Intron | 238 - 639 | 55639562-55639963 | 402 | sequence | - |
| 2 | Exon | 640 - 799 | 55639402-55639561 | 160 | sequence | download |
| 2 | Intron | 800 - 19734 | 55620467-55639401 | 18935 | sequence | - |
| 3 | Exon | 19735 - 19855 | 55620346-55620466 | 121 | sequence | download |
| 3 | Intron | 19856 - 33847 | 55606354-55620345 | 13992 | sequence | - |
| 4 | Exon | 33848 - 33895 | 55606306-55606353 | 48 | sequence | download |
| 4 | Intron | 33896 - 34813 | 55605388-55606305 | 918 | sequence | - |
| 5 | Exon | 34814 - 35007 | 55605194-55605387 | 194 | sequence | download |
| 5 | Intron | 35008 - 51377 | 55588824-55605193 | 16370 | sequence | - |
| 6 | Exon | 51378 - 51436 | 55588765-55588823 | 59 | sequence | download |
| 6 | Intron | 51437 - 74817 | 55565384-55588764 | 23381 | sequence | - |
| 7 | Exon | 74818 - 74895 | 55565306-55565383 | 78 | sequence | download |
| 7 | Intron | 74896 - 80089 | 55560112-55565305 | 5194 | sequence | - |
| 8 | Exon | 80090 - 80226 | 55559975-55560111 | 137 | sequence | download |
| 8 | Intron | 80227 - 99462 | 55540739-55559974 | 19236 | sequence | - |
| 9 | Exon | 99463 - 101894 | 55538307-55540738 | 2432 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-107 | 99745 - 99752 | 55540449 - 55540456 | pictar |
| M2 | hsa-miR-103 | 99745 - 99752 | 55540449 - 55540456 | pictar |
| M3 | hsa-miR-1185 | 99770 - 99776 | 55540425 - 55540431 | pita |
| M4 | hsa-miR-636 | 99780 - 99787 | 55540414 - 55540421 | pita |
| M5 | hsa-miR-218 | 99781 - 99788 | 55540413 - 55540420 | pita |
| M6 | hsa-miR-218 | 99781 - 99788 | 55540413 - 55540420 | pictar |
| M7 | hsa-miR-218 | 99781 - 99788 | 55540413 - 55540420 | targetscan |
| M8 | hsa-miR-107 | 99844 - 99851 | 55540350 - 55540357 | pictar |
| M9 | hsa-miR-103 | 99844 - 99851 | 55540350 - 55540357 | pictar |
| M10 | hsa-miR-107 | 99847 - 99854 | 55540347 - 55540354 | pictar |
| M11 | hsa-miR-103 | 99847 - 99854 | 55540347 - 55540354 | pictar |
| Group 2 | ||||
| M1 | hsa-miR-30e-5p | 100446 - 100453 | 55539748 - 55539755 | pictar |
| Group 3 | ||||
| M1 | hsa-miR-107 | 100953 - 100960 | 55539241 - 55539248 | pictar |
| M2 | hsa-miR-218 | 101219 - 101226 | 55538975 - 55538982 | pictar |
| M3 | hsa-miR-103 | 101227 - 101234 | 55538967 - 55538974 | pictar |
| M4 | hsa-miR-103 | 101227 - 101234 | 55538967 - 55538974 | targetscan |
| M5 | hsa-miR-107 | 101227 - 101234 | 55538967 - 55538974 | targetscan |
| M6 | hsa-miR-107 | 101227 - 101234 | 55538967 - 55538974 | pictar |
| M7 | hsa-miR-107 | 101290 - 101297 | 55538904 - 55538911 | pictar |
| M8 | hsa-miR-103 | 101290 - 101297 | 55538904 - 55538911 | pictar |
| M9 | hsa-miR-218 | 101310 - 101330 | 55538871 - 55538891 | miranda |
| M10 | hsa-miR-218 | 101322 - 101329 | 55538872 - 55538879 | pictar |
| M11 | hsa-miR-218 | 101322 - 101330 | 55538871 - 55538879 | targetscan |
| M12 | hsa-miR-218 | 101323 - 101329 | 55538872 - 55538878 | pita |
| M13 | hsa-miR-30e | 101337 - 101345 | 55538856 - 55538864 | targetscan |
| M14 | hsa-miR-30a | 101337 - 101345 | 55538856 - 55538864 | targetscan |
| M15 | hsa-miR-30b | 101337 - 101345 | 55538856 - 55538864 | targetscan |
| M16 | hsa-miR-30c | 101337 - 101345 | 55538856 - 55538864 | targetscan |
| M17 | hsa-miR-30d | 101337 - 101345 | 55538856 - 55538864 | targetscan |
| M18 | hsa-miR-30a-5p | 101338 - 101345 | 55538856 - 55538863 | pictar |
| M19 | hsa-miR-30e-5p | 101338 - 101345 | 55538856 - 55538863 | pictar |
| M20 | hsa-miR-30d | 101338 - 101345 | 55538856 - 55538863 | pictar |
| M21 | hsa-miR-30c | 101338 - 101345 | 55538856 - 55538863 | pictar |
| M22 | hsa-miR-30b | 101338 - 101345 | 55538856 - 55538863 | pictar |
| M23 | hsa-miR-218 | 101432 - 101439 | 55538762 - 55538769 | pictar |
| M24 | hsa-miR-218 | 101432 - 101439 | 55538762 - 55538769 | targetscan |
| M25 | hsa-miR-342-3p | 101562 - 101584 | 55538617 - 55538639 | miranda |
| M26 | hsa-miR-103 | 101794 - 101801 | 55538400 - 55538407 | pictar |
| M27 | hsa-miR-507 | 101870 - 101876 | 55538325 - 55538331 | pita |
| M28 | hsa-miR-557 | 101870 - 101876 | 55538325 - 55538331 | pita |
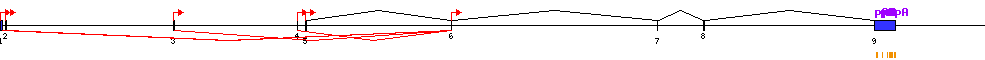















Please wait while loading ...