
| Gene |  |
||
|---|---|---|---|
| Symbol | SEPT14 | ||
| Synonyms | FLJ44060 | ||
| Description | septin 14 | ||
| EnsEMBL link | ENSG00000154997 | ||
| Chromosome | 7 - reverse strand | ||
| Chromosomal location (UCSC link) |
55861237 - 55930482 (69246bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_207366 AK301928 AK126048 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 55930482 | NM_207366 |
| P2 | 1 | 38 | 55930445 | AK301928 |
| P3 | 6 | 28241 | 55902242 | AK126048 |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 10 | 67305 | 55863178 | no | AK301928 | Full_length | |
| PA2 | 10 | 69246 | 55861237 | AATAAA | no | NM_207366 | Cdna |
| PA3 | 10 | 69246 | 55861237 | AATAAA | no | AK126048 | Full_length |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 20640 | 3 | 4 |
| 20640 | 4 | 5 |
| 20640 | 5 | 6 |
| 20640 | 6 | 8 |
| 20640 | 7 | 9 |
| 20640 | 8 | 10 |
| 20640 | 9 | 11 |
| 20640 | 10 | 12 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 2 |
| 6 | 1 |
| Transcription termination & last exon(s) | |
| 10 | 3 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 102 | 55930381-55930482 | 102 | sequence | download |
| 1 | Intron | 103 - 778 | 55929705-55930380 | 676 | sequence | - |
| 2 | Exon | 779 - 847 | 55929636-55929704 | 69 | sequence | download |
| 2 | Intron | 848 - 16152 | 55914331-55929635 | 15305 | sequence | - |
| 3 | Exon | 16153 - 16273 | 55914210-55914330 | 121 | sequence | download |
| 3 | Intron | 16274 - 18071 | 55912412-55914209 | 1798 | sequence | - |
| 4 | Exon | 18072 - 18267 | 55912216-55912411 | 196 | sequence | download |
| 4 | Intron | 18268 - 19661 | 55910822-55912215 | 1394 | sequence | - |
| 5 | Exon | 19662 - 19848 | 55910635-55910821 | 187 | sequence | download |
| 5 | Intron | 19849 - 28203 | 55902280-55910634 | 8355 | sequence | - |
| 6 | Exon | 28204 - 28365 | 55902118-55902279 | 162 | sequence | download |
| 6 | Intron | 28366 - 43566 | 55886917-55902117 | 15201 | sequence | - |
| 7 | Exon | 43567 - 43663 | 55886820-55886916 | 97 | sequence | download |
| 7 | Intron | 43664 - 55531 | 55874952-55886819 | 11868 | sequence | - |
| 8 | Exon | 55532 - 55700 | 55874783-55874951 | 169 | sequence | download |
| 8 | Intron | 55701 - 57399 | 55873084-55874782 | 1699 | sequence | - |
| 9 | Exon | 57400 - 57532 | 55872951-55873083 | 133 | sequence | download |
| 9 | Intron | 57533 - 66697 | 55863786-55872950 | 9165 | sequence | - |
| 10 | Exon | 66698 - 69246 | 55861237-55863785 | 2549 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-146a | 66943 - 66967 | 55863516 - 55863540 | miranda |
| M2 | hsa-miR-146b-5p | 66943 - 66967 | 55863516 - 55863540 | miranda |
| Group 2 | ||||
| M1 | hsa-miR-182 | 67407 - 67435 | 55863048 - 55863076 | miranda |
| M2 | hsa-miR-105 | 67768 - 67774 | 55862709 - 55862715 | pita |
| M3 | hsa-miR-607 | 67770 - 67776 | 55862707 - 55862713 | pita |
| M4 | hsa-miR-374b | 68689 - 68711 | 55861772 - 55861794 | miranda |
| M5 | hsa-miR-374a | 68689 - 68711 | 55861772 - 55861794 | miranda |
| M6 | hsa-miR-340 | 68775 - 68796 | 55861687 - 55861708 | miranda |
| M7 | hsa-miR-422a | 68802 - 68824 | 55861659 - 55861681 | miranda |
| M8 | hsa-miR-222 | 68835 - 68859 | 55861624 - 55861648 | miranda |
| M9 | hsa-miR-221 | 68836 - 68859 | 55861624 - 55861647 | miranda |
| M10 | hsa-miR-221 | 68851 - 68859 | 55861624 - 55861632 | targetscan |
| M11 | hsa-miR-222 | 68851 - 68859 | 55861624 - 55861632 | targetscan |
| M12 | hsa-miR-377 | 68930 - 68952 | 55861531 - 55861553 | miranda |
| M13 | hsa-miR-376c | 68977 - 68997 | 55861486 - 55861506 | miranda |
| M14 | hsa-miR-26a | 68999 - 69021 | 55861462 - 55861484 | miranda |
| M15 | hsa-miR-26b | 69000 - 69021 | 55861462 - 55861483 | miranda |

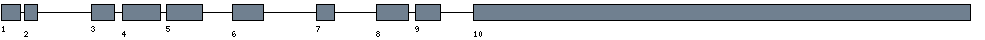




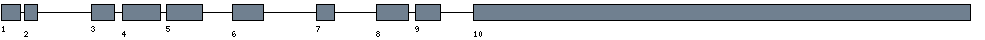
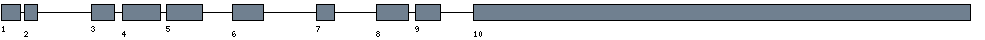

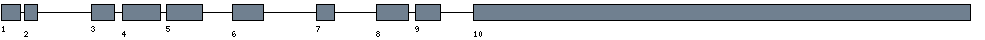

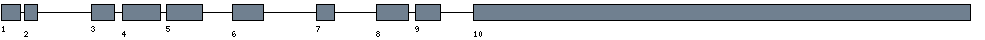

Please wait while loading ...