
| Gene |  |
||
|---|---|---|---|
| Symbol | TEX2 | ||
| Synonyms | HT008, KIAA1738, TMEM96 | ||
| Description | testis expressed 2 | ||
| EnsEMBL link | ENSG00000136478 | ||
| Chromosome | 17 - reverse strand | ||
| Chromosomal location (UCSC link) |
62224796 - 62340656 (115861bp) | ||
| Sequence | sequence | ||
| Associated transcripts | BC040522 BC036672 NM_018469 AK091214 AL834251 BC040521 AL832371 CR627433 AB051525 BC008896 AK091156 AK000758 BC108279 AF163260 BC033661 AF220182 AK000369 AK314316 BX648260 AK130183 |
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 6 | 21 | BC040522 BC036672 AK091214 AL834251 AL832371 CR627433 AB051525 BC008896 AK091156 AK000758 BC108279 AF163260 BC033661 |
|
| Intron retentions |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| IR1 | 12 | BX648260 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 62340656 | BC040522 BC036672 |
| P2 | 1 | 56 | 62340601 | AK091214 |
| P3 | 2 | 32540 | 62308117 | CR627433 |
| P4 | 3 | 49910 | 62290747 | AK091156 |
| P5 | 3 | 50663 | 62289994 | AK000758 |
| P6 | 7 | 85895 | 62254762 | AK000369 |
| P7 | 7 | 85950 | 62254707 | AK314316 |
| P8 | 10 | 107063 | 62233594 | BX648260 |
| P9 | 13 | 114491 | 62226166 | AK130183 |
| P10 | 13 | 115622 | 62225035 | AK130183 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| BC040522 | sequence |
| BC036672 | sequence |
| NM_018469 | sequence |
| AK091214 | sequence |
| AL834251 | sequence |
| BC040521 | sequence |
| AL832371 | sequence |
| CR627433 | sequence |
| AB051525 | sequence |
| BC008896 | sequence |
| AK091156 | sequence |
| AK000758 | sequence |
| BC108279 | sequence |
| AF163260 | sequence |
| BC033661 | sequence |
| AF220182 | sequence |
| AK000369 | sequence |
| AK314316 | sequence |
| BX648260 | sequence |
| AK130183 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 11 | 110239 | 62230418 | no | AK091214 | Full_length | |
| PA2 | 13 | 114328 | 62226329 | no | AK314316 | Full_length | |
| PA3 | 13 | 115607 | 62225050 | ATTAAA | no | AK091156 | Full_length |
| PA4 | 13 | 115607 | 62225050 | ATTAAA | no | AK000758 | Full_length |
| PA5 | 13 | 115607 | 62225050 | ATTAAA | yes | AK000369 | Full_length |
| PA6 | 13 | 115861 | 62224796 | AATAAA | yes | NM_018469 | Cdna |
| PA7 | 13 | 115861 | 62224796 | AATAAA | yes | BC040521 | Cdna |
| PA8 | 13 | 115861 | 62224796 | AATAAA | yes | BC108279 | Cdna |
| PA9 | 13 | 115861 | 62224796 | AATAAA | no | AK130183 | Full_length |
| PA10 | 13 | 117305 | 62223352 | AATACA | no | AI420534 | 3prime_est |
| Exon skipping |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ES1 | 2 | BC040522 BC036672 NM_018469 AK091214 AL834251 BC040521 AL832371 |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 19072 | 1 | 1 |
| 19072 | 3 | 3 |
| 19072 | 4 | 4 |
| 19072 | 5 | 5 |
| 19072 | 6 | 6 |
| 19072 | 7 | 7 |
| 19072 | 8 | 8 |
| 19072 | 9 | 9 |
| 19072 | 10 | 10 |
| 19072 | 11 | 11 |
| 19072 | 12 | 12 |
| 19072 | 13 | 13 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 2 |
| 2 | 1 |
| 3 | 2 |
| 7 | 2 |
| 10 | 1 |
| 13 | 2 |
| Transcription termination & last exon(s) | |
| 11 | 1 |
| 13 | 9 |
| Exon skipping | |
| 2 | 7 |
| Alternative 3' splice sites | |
| 6 | 1 |
| Intron retentions | |
| 12 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 129 | 62340528-62340656 | 129 | sequence | download |
| 1 | Intron | 130 - 32539 | 62308118-62340527 | 32410 | sequence | - |
| 2 | Exon | 32540 - 32688 | 62307969-62308117 | 149 | sequence | download |
| 2 | Intron | 32689 - 49054 | 62291603-62307968 | 16366 | sequence | - |
| 3 | Exon | 49055 - 50723 | 62289934-62291602 | 1669 | sequence | download |
| 3 | Intron | 50724 - 68201 | 62272456-62289933 | 17478 | sequence | - |
| 4 | Exon | 68202 - 68402 | 62272255-62272455 | 201 | sequence | download |
| 4 | Intron | 68403 - 69407 | 62271250-62272254 | 1005 | sequence | - |
| 5 | Exon | 69408 - 69738 | 62270919-62271249 | 331 | sequence | download |
| 5 | Intron | 69739 - 74860 | 62265797-62270918 | 5122 | sequence | - |
| 6 | Exon | 74861 - 75129 | 62265528-62265796 | 269 | sequence | download |
| 6 | Intron | 75130 - 85825 | 62254832-62265527 | 10696 | sequence | - |
| 7 | Exon | 85826 - 85972 | 62254685-62254831 | 147 | sequence | download |
| 7 | Intron | 85973 - 92097 | 62248560-62254684 | 6125 | sequence | - |
| 8 | Exon | 92098 - 92197 | 62248460-62248559 | 100 | sequence | download |
| 8 | Intron | 92198 - 102363 | 62238294-62248459 | 10166 | sequence | - |
| 9 | Exon | 102364 - 102496 | 62238161-62238293 | 133 | sequence | download |
| 9 | Intron | 102497 - 108329 | 62232328-62238160 | 5833 | sequence | - |
| 10 | Exon | 108330 - 108455 | 62232202-62232327 | 126 | sequence | download |
| 10 | Intron | 108456 - 110142 | 62230515-62232201 | 1687 | sequence | - |
| 11 | Exon | 110143 - 110352 | 62230305-62230514 | 210 | sequence | download |
| 11 | Intron | 110353 - 112335 | 62228322-62230304 | 1983 | sequence | - |
| 12 | Exon | 112336 - 112456 | 62228201-62228321 | 121 | sequence | download |
| 12 | Intron | 112457 - 114205 | 62226452-62228200 | 1749 | sequence | - |
| 13 | Exon | 114206 - 115861 | 62224796-62226451 | 1656 | sequence | download |










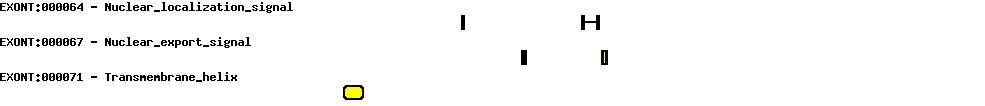



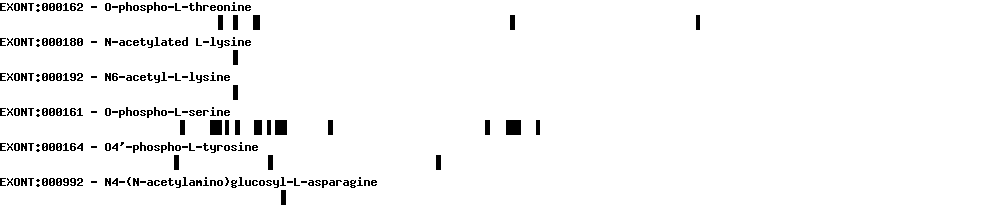


Please wait while loading ...