
| Gene |  |
||
|---|---|---|---|
| Symbol | ERN1 | ||
| Synonyms | IRE1, IRE1P | ||
| Description | endoplasmic reticulum to nucleus signaling 1 | ||
| EnsEMBL link | ENSG00000178607 | ||
| Chromosome | 17 - reverse strand | ||
| Chromosomal location (UCSC link) |
62120353 - 62207528 (87176bp) | ||
| Sequence | sequence | ||
| Associated transcripts | AK292403 NM_001433 AB209869 AF059198 BC130405 BC130407 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 62207528 | AK292403 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| AK292403 | sequence |
| NM_001433 | sequence |
| AB209869 | sequence |
| AF059198 | sequence |
| BC130405 | sequence |
| BC130407 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 22 | 86219 | 62121310 | no | AK292403 | Full_length | |
| PA2 | 22 | 87176 | 62120353 | no | AB209869 | Cdna | |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 1 |
| Transcription termination & last exon(s) | |
| 22 | 2 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| 16 | Expression | Expression | Expression |
| 17 | Expression | Expression | Expression |
| 18 | Expression | Expression | Expression |
| 19 | Expression | Expression | Expression |
| 20 | Expression | Expression | Expression |
| 21 | Expression | Expression | Expression |
| 22 | Expression | Expression | Expression |
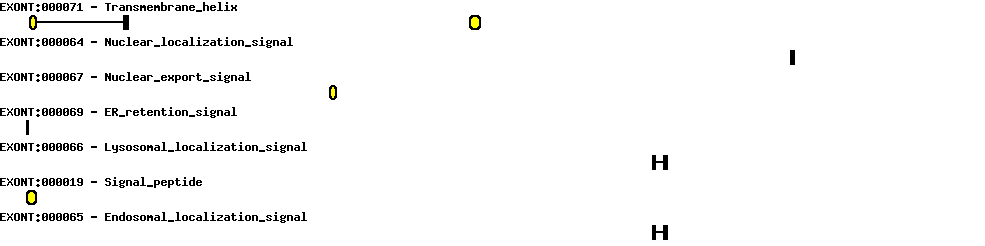
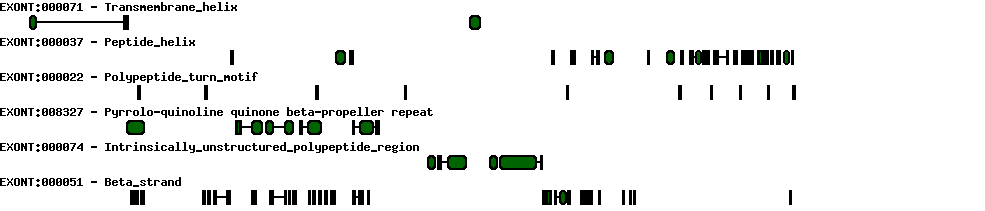
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 193 | 62207336-62207528 | 193 | sequence | download |
| 1 | Intron | 194 - 31927 | 62175602-62207335 | 31734 | sequence | - |
| 2 | Exon | 31928 - 32048 | 62175481-62175601 | 121 | sequence | download |
| 2 | Intron | 32049 - 49360 | 62158169-62175480 | 17312 | sequence | - |
| 3 | Exon | 49361 - 49394 | 62158135-62158168 | 34 | sequence | download |
| 3 | Intron | 49395 - 50434 | 62157095-62158134 | 1040 | sequence | - |
| 4 | Exon | 50435 - 50507 | 62157022-62157094 | 73 | sequence | download |
| 4 | Intron | 50508 - 54921 | 62152608-62157021 | 4414 | sequence | - |
| 5 | Exon | 54922 - 54994 | 62152535-62152607 | 73 | sequence | download |
| 5 | Intron | 54995 - 58065 | 62149464-62152534 | 3071 | sequence | - |
| 6 | Exon | 58066 - 58188 | 62149341-62149463 | 123 | sequence | download |
| 6 | Intron | 58189 - 61877 | 62145652-62149340 | 3689 | sequence | - |
| 7 | Exon | 61878 - 61979 | 62145550-62145651 | 102 | sequence | download |
| 7 | Intron | 61980 - 63236 | 62144293-62145549 | 1257 | sequence | - |
| 8 | Exon | 63237 - 63498 | 62144031-62144292 | 262 | sequence | download |
| 8 | Intron | 63499 - 64881 | 62142648-62144030 | 1383 | sequence | - |
| 9 | Exon | 64882 - 64960 | 62142569-62142647 | 79 | sequence | download |
| 9 | Intron | 64961 - 66017 | 62141512-62142568 | 1057 | sequence | - |
| 10 | Exon | 66018 - 66183 | 62141346-62141511 | 166 | sequence | download |
| 10 | Intron | 66184 - 69581 | 62137948-62141345 | 3398 | sequence | - |
| 11 | Exon | 69582 - 69700 | 62137829-62137947 | 119 | sequence | download |
| 11 | Intron | 69701 - 72175 | 62135354-62137828 | 2475 | sequence | - |
| 12 | Exon | 72176 - 72367 | 62135162-62135353 | 192 | sequence | download |
| 12 | Intron | 72368 - 74220 | 62133309-62135161 | 1853 | sequence | - |
| 13 | Exon | 74221 - 74494 | 62133035-62133308 | 274 | sequence | download |
| 13 | Intron | 74495 - 75340 | 62132189-62133034 | 846 | sequence | - |
| 14 | Exon | 75341 - 75431 | 62132098-62132188 | 91 | sequence | download |
| 14 | Intron | 75432 - 75729 | 62131800-62132097 | 298 | sequence | - |
| 15 | Exon | 75730 - 75919 | 62131610-62131799 | 190 | sequence | download |
| 15 | Intron | 75920 - 76797 | 62130732-62131609 | 878 | sequence | - |
| 16 | Exon | 76798 - 76897 | 62130632-62130731 | 100 | sequence | download |
| 16 | Intron | 76898 - 77189 | 62130340-62130631 | 292 | sequence | - |
| 17 | Exon | 77190 - 77389 | 62130140-62130339 | 200 | sequence | download |
| 17 | Intron | 77390 - 80966 | 62126563-62130139 | 3577 | sequence | - |
| 18 | Exon | 80967 - 81114 | 62126415-62126562 | 148 | sequence | download |
| 18 | Intron | 81115 - 82183 | 62125346-62126414 | 1069 | sequence | - |
| 19 | Exon | 82184 - 82311 | 62125218-62125345 | 128 | sequence | download |
| 19 | Intron | 82312 - 84686 | 62122843-62125217 | 2375 | sequence | - |
| 20 | Exon | 84687 - 84810 | 62122719-62122842 | 124 | sequence | download |
| 20 | Intron | 84811 - 85241 | 62122288-62122718 | 431 | sequence | - |
| 21 | Exon | 85242 - 85309 | 62122220-62122287 | 68 | sequence | download |
| 21 | Intron | 85310 - 85968 | 62121561-62122219 | 659 | sequence | - |
| 22 | Exon | 85969 - 87176 | 62120353-62121560 | 1208 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-124 | 86293 - 86300 | 62121229 - 62121236 | targetscan |
| M2 | hsa-miR-506 | 86293 - 86300 | 62121229 - 62121236 | targetscan |
| M3 | hsa-miR-124 | 86294 - 86300 | 62121229 - 62121235 | pita |
| M4 | hsa-miR-506 | 86294 - 86300 | 62121229 - 62121235 | pita |
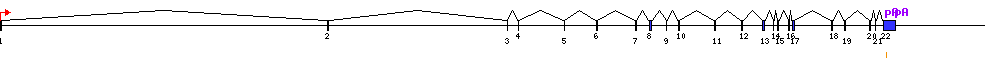
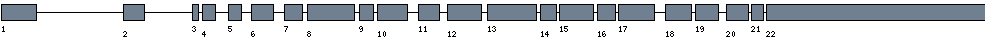
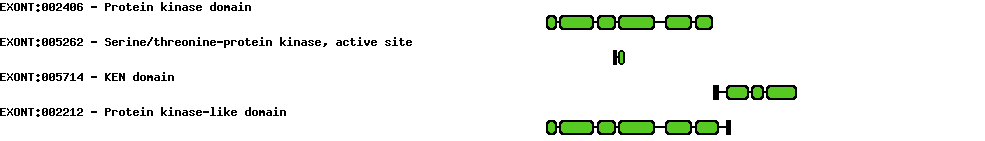
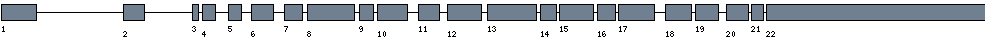

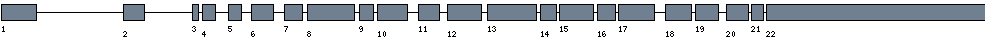

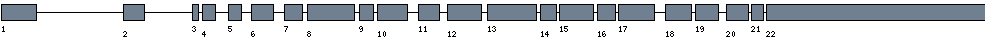
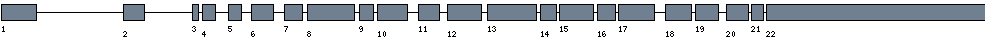

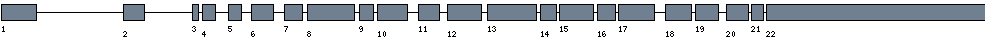
Please wait while loading ...