
 FasterDB identifier: 17164 - POLG2 (HP55, MTPOLB)
polymerase (DNA directed), gamma 2, accessory subunit
FasterDB identifier: 17164 - POLG2 (HP55, MTPOLB)
polymerase (DNA directed), gamma 2, accessory subunit

| Gene |  |
||
|---|---|---|---|
| Symbol | POLG2 | ||
| Synonyms | HP55, MTPOLB | ||
| Description | polymerase (DNA directed), gamma 2, accessory subunit | ||
| EnsEMBL link | ENSG00000136480 | ||
| Chromosome | 17 - reverse strand | ||
| Chromosomal location (UCSC link) |
62473902 - 62493184 (19283bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_007215 BC009194 U94703 AF184344 AF142992 AK307801 AF177201 BC000913 CR617858 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 62493184 | NM_007215 |
| P2 | 1 | 48 | 62493137 | AK307801 |
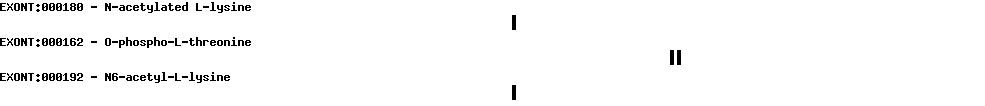
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_007215 | sequence |
| BC009194 | sequence |
| U94703 | sequence |
| AF184344 | sequence |
| AF142992 | sequence |
| AK307801 | sequence |
| AF177201 | sequence |
| BC000913 | sequence |
| CR617858 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 5 | 11474 | 62481711 | no | AK307801 | Full_length | |
| PA2 | 8 | 19283 | 62473902 | AATAAA | no | CR617858 | Cdna |
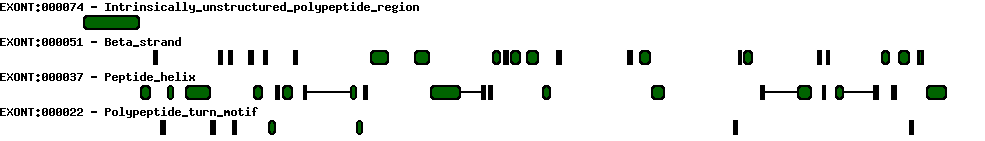
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 19190 | 1 | 1 |
| 19190 | 2 | 2 |
| 19190 | 3 | 3 |
| 19190 | 4 | 4 |
| 19190 | 5 | 5 |
| 19190 | 6 | 6 |
| 19190 | 7 | 7 |
| 19190 | 8 | 9 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 2 |
| Transcription termination & last exon(s) | |
| 5 | 1 |
| 8 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 660 | 62492525-62493184 | 660 | sequence | download |
| 1 | Intron | 661 - 4046 | 62489139-62492524 | 3386 | sequence | - |
| 2 | Exon | 4047 - 4173 | 62489012-62489138 | 127 | sequence | download |
| 2 | Intron | 4174 - 4295 | 62488890-62489011 | 122 | sequence | - |
| 3 | Exon | 4296 - 4401 | 62488784-62488889 | 106 | sequence | download |
| 3 | Intron | 4402 - 6098 | 62487087-62488783 | 1697 | sequence | - |
| 4 | Exon | 6099 - 6272 | 62486913-62487086 | 174 | sequence | download |
| 4 | Intron | 6273 - 11199 | 62481986-62486912 | 4927 | sequence | - |
| 5 | Exon | 11200 - 11340 | 62481845-62481985 | 141 | sequence | download |
| 5 | Intron | 11341 - 14068 | 62479117-62481844 | 2728 | sequence | - |
| 6 | Exon | 14069 - 14149 | 62479036-62479116 | 81 | sequence | download |
| 6 | Intron | 14150 - 16678 | 62476507-62479035 | 2529 | sequence | - |
| 7 | Exon | 16679 - 16779 | 62476406-62476506 | 101 | sequence | download |
| 7 | Intron | 16780 - 19079 | 62474106-62476405 | 2300 | sequence | - |
| 8 | Exon | 19080 - 19283 | 62473902-62474105 | 204 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-185 | 19260 - 19281 | 62473904 - 62473925 | miranda |















Please wait while loading ...