
| Gene |  |
||
|---|---|---|---|
| Symbol | DDX5 | ||
| Synonyms | G17P1, HLR1, p68 | ||
| Description | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | ||
| EnsEMBL link | ENSG00000108654 | ||
| Chromosome | 17 - reverse strand | ||
| Chromosomal location (UCSC link) |
62494374 - 62502484 (8111bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_004396 X52104 BX571764 AK297753 AK297905 AK297192 X15729 AB209919 BC016027 AB451257 BT006943 AB451382 AK310318 CR607012 AK055995 |
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 6 | 52 | AK297192 |
|
| Alternative 5' splice sites (donor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 4 | -51 | AK297192 |
|
| ASS2 | 3 | -22 | NM_004396 X52104 BX571764 AK297753 AK297192 X15729 AB209919 BC016027 AB451257 BT006943 AB451382 AK310318 |
|
| Exon deletions |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ED1 | 3 | AK297753 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 62502484 | NM_004396 |
| P2 | 1 | 78 | 62502407 | AK297753 AK297905 |
| P3 | 1 | 79 | 62502406 | AK297192 |
| P4 | 2 | 534 | 62501951 | AK310318 |
| P5 | 13 | 4878 | 62497607 | AK055995 |
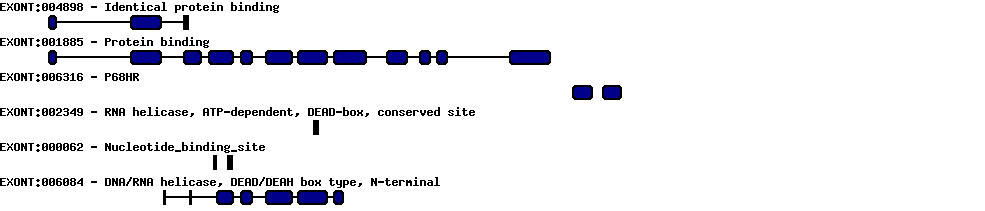
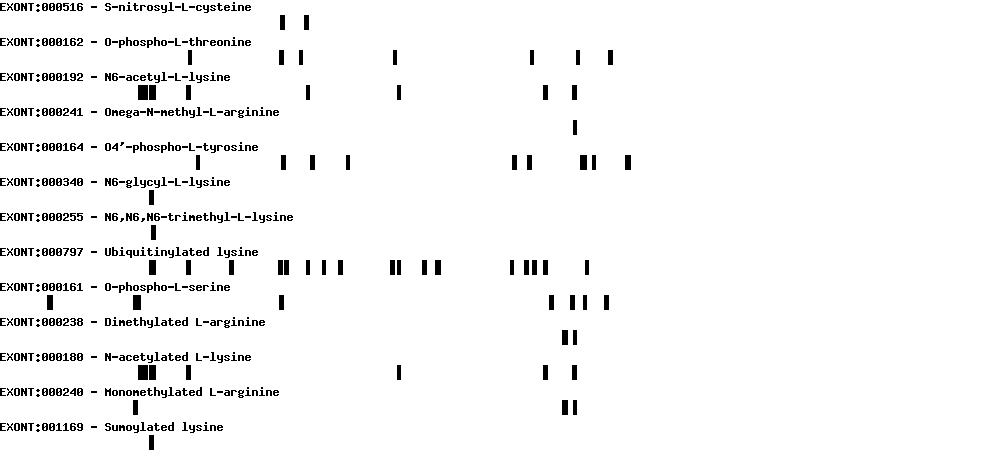
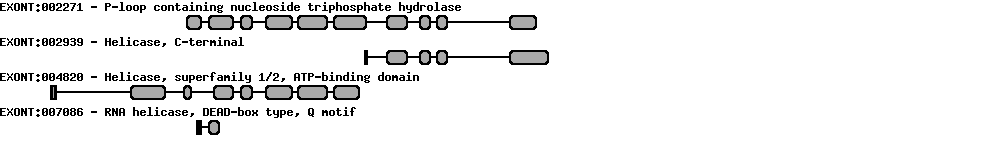
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_004396 | sequence |
| X52104 | sequence |
| BX571764 | sequence |
| AK297753 | sequence |
| AK297905 | sequence |
| AK297192 | sequence |
| X15729 | sequence |
| AB209919 | sequence |
| BC016027 | sequence |
| AB451257 | sequence |
| BT006943 | sequence |
| AB451382 | sequence |
| AK310318 | sequence |
| CR607012 | sequence |
| AK055995 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 14 | 6453 | 62496032 | no | AK310318 | Full_length | |
| PA2 | 14 | 6576 | 62495909 | no | AK297753 | Full_length | |
| PA3 | 14 | 6745 | 62495740 | AATAAA | no | AK297192 | Full_length |
| PA4 | 14 | 6745 | 62495740 | AATAAA | no | AK055995 | Full_length |
| PA5 | 14 | 8111 | 62494374 | AATAAA | yes | NM_004396 | Cdna |
| Exon skipping |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ES1 | 2 | NM_004396 X52104 BX571764 AK297753 AK297905 AK297192 X15729 AB209919 BC016027 AB451257 BT006943 AB451382 |
| ES2 | 4 | AK297905 |
| ES3 | 5 | AK297905 AK297192 |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 19266 | 1 | 2 |
| 19266 | 3 | 3 |
| 19266 | 4 | 4 |
| 19266 | 5 | 5 |
| 19266 | 6 | 6 |
| 19266 | 7 | 7 |
| 19266 | 8 | 8 |
| 19266 | 9 | 9 |
| 19266 | 10 | 10 |
| 19266 | 11 | 11 |
| 19266 | 12 | 12 |
| 19266 | 13 | 13 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 3 |
| 2 | 1 |
| 13 | 1 |
| Transcription termination & last exon(s) | |
| 14 | 5 |
| Exon skipping | |
| 2 | 12 |
| 4 | 1 |
| 5 | 2 |
| Alternative 3' splice sites | |
| 6 | 1 |
| Alternative 5' splice sites | |
| 3 | 1 |
| 4 | 1 |
| Exon deletions | |
| 3 | 1 |
| Intron retentions | |
| 12 | 2 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 291 | 62502194-62502484 | 291 | sequence | download |
| 1 | Intron | 292 - 533 | 62501952-62502193 | 242 | sequence | - |
| 2 | Exon | 534 - 621 | 62501864-62501951 | 88 | sequence | download |
| 2 | Intron | 622 - 1524 | 62500961-62501863 | 903 | sequence | - |
| 3 | Exon | 1525 - 1712 | 62500773-62500960 | 188 | sequence | download |
| 3 | Intron | 1713 - 2048 | 62500437-62500772 | 336 | sequence | - |
| 4 | Exon | 2049 - 2145 | 62500340-62500436 | 97 | sequence | download |
| 4 | Intron | 2146 - 2250 | 62500235-62500339 | 105 | sequence | - |
| 5 | Exon | 2251 - 2384 | 62500101-62500234 | 134 | sequence | download |
| 5 | Intron | 2385 - 2498 | 62499987-62500100 | 114 | sequence | - |
| 6 | Exon | 2499 - 2564 | 62499921-62499986 | 66 | sequence | download |
| 6 | Intron | 2565 - 2795 | 62499690-62499920 | 231 | sequence | - |
| 7 | Exon | 2796 - 2937 | 62499548-62499689 | 142 | sequence | download |
| 7 | Intron | 2938 - 3018 | 62499467-62499547 | 81 | sequence | - |
| 8 | Exon | 3019 - 3179 | 62499306-62499466 | 161 | sequence | download |
| 8 | Intron | 3180 - 3268 | 62499217-62499305 | 89 | sequence | - |
| 9 | Exon | 3269 - 3441 | 62499044-62499216 | 173 | sequence | download |
| 9 | Intron | 3442 - 3817 | 62498668-62499043 | 376 | sequence | - |
| 10 | Exon | 3818 - 3928 | 62498557-62498667 | 111 | sequence | download |
| 10 | Intron | 3929 - 4143 | 62498342-62498556 | 215 | sequence | - |
| 11 | Exon | 4144 - 4205 | 62498280-62498341 | 62 | sequence | download |
| 11 | Intron | 4206 - 4297 | 62498188-62498279 | 92 | sequence | - |
| 12 | Exon | 4298 - 4357 | 62498128-62498187 | 60 | sequence | download |
| 12 | Intron | 4358 - 5593 | 62496892-62498127 | 1236 | sequence | - |
| 13 | Exon | 5594 - 5818 | 62496667-62496891 | 225 | sequence | download |
| 13 | Intron | 5819 - 6040 | 62496445-62496666 | 222 | sequence | - |
| 14 | Exon | 6041 - 8111 | 62494374-62496444 | 2071 | sequence | download |














Please wait while loading ...