
| Gene |  |
||
|---|---|---|---|
| Symbol | CCDC45 | ||
| Synonyms | DKFZp667E1824 | ||
| Description | coiled-coil domain containing 45 | ||
| EnsEMBL link | ENSG00000141325 | ||
| Chromosome | 17 - forward strand | ||
| Chromosomal location (UCSC link) |
62502880 - 62534067 (31188bp) | ||
| Sequence | sequence | ||
| Associated transcripts | AK057326 AK316257 AK290587 AK297409 BC009518 NM_138363 AL832822 BX641136 BC029461 |
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 4 | 19 | AK057326 AK316257 AK290587 AK297409 BC009518 NM_138363 |
|
| Alternative 5' splice sites (donor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 13 | -4 | AK057326 AK290587 AK297409 BC009518 NM_138363 |
|
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 62502880 | AK057326 |
| P2 | 1 | 231 | 62503110 | AK316257 |
| P3 | 1 | 235 | 62503114 | AK290587 |
| P4 | 1 | 274 | 62503153 | AK297409 |
| P5 | 17 | 27255 | 62530134 | BX641136 |
| P6 | 18 | 29371 | 62532250 | BC029461 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| AK057326 | sequence |
| AK316257 | sequence |
| AK290587 | sequence |
| AK297409 | sequence |
| BC009518 | sequence |
| NM_138363 | sequence |
| AL832822 | sequence |
| BX641136 | sequence |
| BC029461 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 9 | 19024 | 62521903 | no | AK316257 | Full_length | |
| PA2 | 15 | 26225 | 62529104 | no | AK057326 | Full_length | |
| PA3 | 20 | 31188 | 62534067 | AATAAA | yes | BC029461 | Cdna |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 4 |
| 17 | 1 |
| 18 | 1 |
| Transcription termination & last exon(s) | |
| 9 | 1 |
| 15 | 1 |
| 20 | 1 |
| Exon skipping | |
| 5 | 2 |
| Alternative 3' splice sites | |
| 4 | 1 |
| Alternative 5' splice sites | |
| 13 | 1 |
| Intron retentions | |
| 17 | 1 |
| 19 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| 16 | Expression | Expression | Expression |
| 17 | Expression | Expression | Expression |
| 18 | Expression | Expression | Expression |
| 19 | Expression | Expression | Expression |
| 20 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 355 | 62502880 - 62503234 | 355 | sequence | download |
| 1 | Intron | 356 - 1830 | 62503235 - 62504709 | 1475 | sequence | - |
| 2 | Exon | 1831 - 1959 | 62504710 - 62504838 | 129 | sequence | download |
| 2 | Intron | 1960 - 3411 | 62504839 - 62506290 | 1452 | sequence | - |
| 3 | Exon | 3412 - 3519 | 62506291 - 62506398 | 108 | sequence | download |
| 3 | Intron | 3520 - 7467 | 62506399 - 62510346 | 3948 | sequence | - |
| 4 | Exon | 7468 - 7597 | 62510347 - 62510476 | 130 | sequence | download |
| 4 | Intron | 7598 - 9961 | 62510477 - 62512840 | 2364 | sequence | - |
| 5 | Exon | 9962 - 10067 | 62512841 - 62512946 | 106 | sequence | download |
| 5 | Intron | 10068 - 12559 | 62512947 - 62515438 | 2492 | sequence | - |
| 6 | Exon | 12560 - 12675 | 62515439 - 62515554 | 116 | sequence | download |
| 6 | Intron | 12676 - 14640 | 62515555 - 62517519 | 1965 | sequence | - |
| 7 | Exon | 14641 - 14766 | 62517520 - 62517645 | 126 | sequence | download |
| 7 | Intron | 14767 - 15940 | 62517646 - 62518819 | 1174 | sequence | - |
| 8 | Exon | 15941 - 16134 | 62518820 - 62519013 | 194 | sequence | download |
| 8 | Intron | 16135 - 19008 | 62519014 - 62521887 | 2874 | sequence | - |
| 9 | Exon | 19009 - 19121 | 62521888 - 62522000 | 113 | sequence | download |
| 9 | Intron | 19122 - 19309 | 62522001 - 62522188 | 188 | sequence | - |
| 10 | Exon | 19310 - 19439 | 62522189 - 62522318 | 130 | sequence | download |
| 10 | Intron | 19440 - 20349 | 62522319 - 62523228 | 910 | sequence | - |
| 11 | Exon | 20350 - 20503 | 62523229 - 62523382 | 154 | sequence | download |
| 11 | Intron | 20504 - 22526 | 62523383 - 62525405 | 2023 | sequence | - |
| 12 | Exon | 22527 - 22666 | 62525406 - 62525545 | 140 | sequence | download |
| 12 | Intron | 22667 - 24164 | 62525546 - 62527043 | 1498 | sequence | - |
| 13 | Exon | 24165 - 24261 | 62527044 - 62527140 | 97 | sequence | download |
| 13 | Intron | 24262 - 25128 | 62527141 - 62528007 | 867 | sequence | - |
| 14 | Exon | 25129 - 25261 | 62528008 - 62528140 | 133 | sequence | download |
| 14 | Intron | 25262 - 26077 | 62528141 - 62528956 | 816 | sequence | - |
| 15 | Exon | 26078 - 26247 | 62528957 - 62529126 | 170 | sequence | download |
| 15 | Intron | 26248 - 26355 | 62529127 - 62529234 | 108 | sequence | - |
| 16 | Exon | 26356 - 26430 | 62529235 - 62529309 | 75 | sequence | download |
| 16 | Intron | 26431 - 27823 | 62529310 - 62530702 | 1393 | sequence | - |
| 17 | Exon | 27824 - 27976 | 62530703 - 62530855 | 153 | sequence | download |
| 17 | Intron | 27977 - 29840 | 62530856 - 62532719 | 1864 | sequence | - |
| 18 | Exon | 29841 - 29987 | 62532720 - 62532866 | 147 | sequence | download |
| 18 | Intron | 29988 - 30279 | 62532867 - 62533158 | 292 | sequence | - |
| 19 | Exon | 30280 - 30351 | 62533159 - 62533230 | 72 | sequence | download |
| 19 | Intron | 30352 - 30841 | 62533231 - 62533720 | 490 | sequence | - |
| 20 | Exon | 30842 - 31188 | 62533721 - 62534067 | 347 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-425 | 1850 - 1872 | 62504729 - 62504751 | miranda |
| M2 | hsa-miR-150 | 1927 - 1948 | 62504806 - 62504827 | miranda |
| Group 2 | ||||
| M1 | hsa-miR-149 | 9994 - 10019 | 62512873 - 62512898 | miranda |
| Group 3 | ||||
| M1 | hsa-miR-485-5p | 19311 - 19334 | 62522190 - 62522213 | miranda |
| M2 | hsa-miR-216b | 19355 - 19376 | 62522234 - 62522255 | miranda |
| M3 | hsa-miR-375 | 19364 - 19386 | 62522243 - 62522265 | miranda |
| Group 4 | ||||
| M1 | hsa-miR-204 | 24170 - 24191 | 62527049 - 62527070 | miranda |
| M2 | hsa-miR-211 | 24170 - 24191 | 62527049 - 62527070 | miranda |
| Group 5 | ||||
| M1 | hsa-miR-876-5p | 26218 - 26240 | 62529097 - 62529119 | miranda |
| Group 6 | ||||
| M1 | hsa-miR-149 | 31019 - 31026 | 62533898 - 62533905 | pictar |
| M2 | hsa-miR-149 | 31067 - 31088 | 62533946 - 62533967 | miranda |
| M3 | hsa-miR-149 | 31070 - 31077 | 62533949 - 62533956 | pictar |
| M4 | hsa-miR-149 | 31082 - 31089 | 62533961 - 62533968 | pictar |
| M5 | hsa-miR-149 | 31099 - 31106 | 62533978 - 62533985 | pictar |














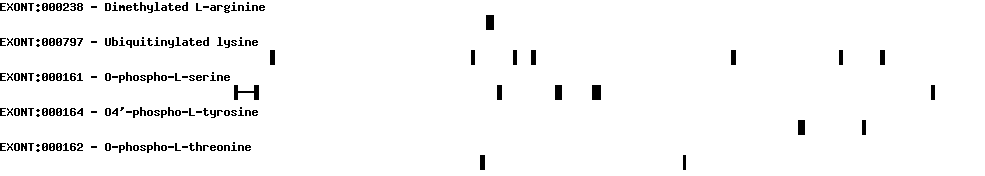


Please wait while loading ...