
 FasterDB identifier: 2271 - KDELR3 (ENSG00000100196)
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
FasterDB identifier: 2271 - KDELR3 (ENSG00000100196)
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3

| Gene |  |
||
|---|---|---|---|
| Symbol | KDELR3 | ||
| Synonyms | ENSG00000100196 | ||
| Description | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 | ||
| EnsEMBL link | ENSG00000100196 | ||
| Chromosome | 22 - forward strand | ||
| Chromosomal location (UCSC link) |
38864067 - 38879487 (15421bp) | ||
| Sequence | sequence | ||
| Associated transcripts | CR590826 CR594497 CR594712 AK292102 NM_006855 AL035082 NM_016657 AK222930 CR612711 BC001277 AK129888 AK223238 AL035081 BC096722 BC098131 BC098156 BC098336 CR600149 CR456509 CR614427 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 38864067 | CR590826 CR594497 CR594712 |
| P2 | 1 | 9 | 38864075 | AK292102 |
| P3 | 1 | 35 | 38864101 | AK129888 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| CR590826 | sequence |
| CR594497 | sequence |
| CR594712 | sequence |
| AK292102 | sequence |
| NM_006855 | sequence |
| AL035082 | sequence |
| NM_016657 | sequence |
| AK222930 | sequence |
| CR612711 | sequence |
| BC001277 | sequence |
| AK129888 | sequence |
| AK223238 | sequence |
| AL035081 | sequence |
| BC096722 | sequence |
| BC098131 | sequence |
| BC098156 | sequence |
| BC098336 | sequence |
| CR600149 | sequence |
| CR456509 | sequence |
| CR614427 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 4 | 13572 | 38877638 | AATAAA | no | AL035082 | Cdna |
| PA2 | 4 | 13572 | 38877638 | AATAAA | no | NM_016657 | Cdna |
| PA3 | 5 | 14874 | 38878940 | ACTAAA | no | AK292102 | Full_length |
| PA4 | 5 | 15421 | 38879487 | no | CR614427 | Cdna | |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 16644 | 1 | 1 |
| 16644 | 2 | 2 |
| 16644 | 3 | 3 |
| 16644 | 4 | 4 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 3 |
| Transcription termination & last exon(s) | |
| 4 | 2 |
| 5 | 2 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 263 | 38864067 - 38864329 | 263 | sequence | download |
| 1 | Intron | 264 - 6461 | 38864330 - 38870527 | 6198 | sequence | - |
| 2 | Exon | 6462 - 6562 | 38870528 - 38870628 | 101 | sequence | download |
| 2 | Intron | 6563 - 11531 | 38870629 - 38875597 | 4969 | sequence | - |
| 3 | Exon | 11532 - 11690 | 38875598 - 38875756 | 159 | sequence | download |
| 3 | Intron | 11691 - 13150 | 38875757 - 38877216 | 1460 | sequence | - |
| 4 | Exon | 13151 - 13403 | 38877217 - 38877469 | 253 | sequence | download |
| 4 | Intron | 13404 - 14434 | 38877470 - 38878500 | 1031 | sequence | - |
| 5 | Exon | 14435 - 15421 | 38878501 - 38879487 | 987 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-153 | 13512 - 13533 | 38877578 - 38877599 | miranda |
| M2 | hsa-miR-544 | 13514 - 13537 | 38877580 - 38877603 | miranda |
| Group 2 | ||||
| M1 | hsa-miR-181d | 14512 - 14534 | 38878578 - 38878600 | miranda |
| M2 | hsa-miR-181a | 14512 - 14534 | 38878578 - 38878600 | miranda |
| M3 | hsa-miR-181b | 14512 - 14534 | 38878578 - 38878600 | miranda |
| M4 | hsa-miR-181c | 14513 - 14534 | 38878579 - 38878600 | miranda |
| Group 3 | ||||
| M1 | hsa-miR-154 | 15100 - 15121 | 38879166 - 38879187 | miranda |
| M2 | hsa-miR-19a | 15125 - 15150 | 38879191 - 38879216 | miranda |
| M3 | hsa-miR-19b | 15128 - 15150 | 38879194 - 38879216 | miranda |
| M4 | hsa-miR-137 | 15152 - 15177 | 38879218 - 38879243 | miranda |
| M5 | hsa-miR-300 | 15220 - 15241 | 38879286 - 38879307 | miranda |
| M6 | hsa-miR-381 | 15220 - 15241 | 38879286 - 38879307 | miranda |
| M7 | hsa-miR-488 | 15238 - 15258 | 38879304 - 38879324 | miranda |
| M8 | hsa-miR-137 | 15343 - 15365 | 38879409 - 38879431 | miranda |
| M9 | hsa-miR-137 | 15358 - 15366 | 38879424 - 38879432 | targetscan |
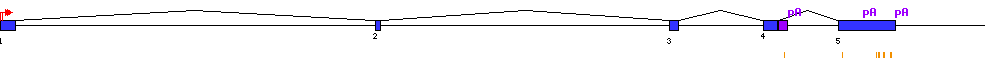
















Please wait while loading ...