
 FasterDB identifier: 2289 - DMC1 (LIM15)
DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast)
FasterDB identifier: 2289 - DMC1 (LIM15)
DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast)

| Gene |  |
||
|---|---|---|---|
| Symbol | DMC1 | ||
| Synonyms | LIM15 | ||
| Description | DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast) | ||
| EnsEMBL link | ENSG00000100206 | ||
| Chromosome | 22 - reverse strand | ||
| Chromosomal location (UCSC link) |
38914954 - 38966201 (51248bp) | ||
| Sequence | sequence | ||
| Associated transcripts | AK292617 BC035658 NM_007068 BC125164 BC125163 D63882 CR456486 D64108 AK297664 |
| Alternative 5' splice sites (donor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 8 | -582 | AK292617 NM_007068 BC125164 BC125163 D63882 CR456486 D64108 |
|
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 38966201 | AK292617 |
| P2 | 2 | 1788 | 38964414 | AK297664 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| AK292617 | sequence |
| BC035658 | sequence |
| NM_007068 | sequence |
| BC125164 | sequence |
| BC125163 | sequence |
| D63882 | sequence |
| CR456486 | sequence |
| D64108 | sequence |
| AK297664 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 9 | 24429 | 38941773 | AATAAA | yes | BC035658 | Cdna |
| PA2 | 15 | 50222 | 38915980 | no | AK297664 | Full_length | |
| PA3 | 15 | 51117 | 38915085 | no | AK292617 | Full_length | |
| PA4 | 15 | 51248 | 38914954 | AATAAA | no | NM_007068 | Cdna |
| PA5 | 15 | 51248 | 38914954 | AATAAA | no | D64108 | Cdna |
| Exon skipping |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ES1 | 8 | AK297664 |
| ES2 | 9 | AK292617 NM_007068 BC125164 BC125163 D63882 CR456486 D64108 AK297664 |
| ES3 | 10 | AK297664 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 1 |
| 2 | 1 |
| Transcription termination & last exon(s) | |
| 9 | 1 |
| 15 | 4 |
| Exon skipping | |
| 8 | 1 |
| 9 | 8 |
| 10 | 1 |
| Alternative 5' splice sites | |
| 8 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 154 | 38966048-38966201 | 154 | sequence | download |
| 1 | Intron | 155 - 1907 | 38964295-38966047 | 1753 | sequence | - |
| 2 | Exon | 1908 - 1991 | 38964211-38964294 | 84 | sequence | download |
| 2 | Intron | 1992 - 2569 | 38963633-38964210 | 578 | sequence | - |
| 3 | Exon | 2570 - 2614 | 38963588-38963632 | 45 | sequence | download |
| 3 | Intron | 2615 - 3460 | 38962742-38963587 | 846 | sequence | - |
| 4 | Exon | 3461 - 3607 | 38962595-38962741 | 147 | sequence | download |
| 4 | Intron | 3608 - 7827 | 38958375-38962594 | 4220 | sequence | - |
| 5 | Exon | 7828 - 7910 | 38958292-38958374 | 83 | sequence | download |
| 5 | Intron | 7911 - 14787 | 38951415-38958291 | 6877 | sequence | - |
| 6 | Exon | 14788 - 14840 | 38951362-38951414 | 53 | sequence | download |
| 6 | Intron | 14841 - 17489 | 38948713-38951361 | 2649 | sequence | - |
| 7 | Exon | 17490 - 17531 | 38948671-38948712 | 42 | sequence | download |
| 7 | Intron | 17532 - 20199 | 38946003-38948670 | 2668 | sequence | - |
| 8 | Exon | 20200 - 20854 | 38945348-38946002 | 655 | sequence | download |
| 8 | Intron | 20855 - 24085 | 38942117-38945347 | 3231 | sequence | - |
| 9 | Exon | 24086 - 24429 | 38941773-38942116 | 344 | sequence | download |
| 9 | Intron | 24430 - 30784 | 38935418-38941772 | 6355 | sequence | - |
| 10 | Exon | 30785 - 30876 | 38935326-38935417 | 92 | sequence | download |
| 10 | Intron | 30877 - 31584 | 38934618-38935325 | 708 | sequence | - |
| 11 | Exon | 31585 - 31658 | 38934544-38934617 | 74 | sequence | download |
| 11 | Intron | 31659 - 31787 | 38934415-38934543 | 129 | sequence | - |
| 12 | Exon | 31788 - 31902 | 38934300-38934414 | 115 | sequence | download |
| 12 | Intron | 31903 - 32544 | 38933658-38934299 | 642 | sequence | - |
| 13 | Exon | 32545 - 32605 | 38933597-38933657 | 61 | sequence | download |
| 13 | Intron | 32606 - 48472 | 38917730-38933596 | 15867 | sequence | - |
| 14 | Exon | 48473 - 48589 | 38917613-38917729 | 117 | sequence | download |
| 14 | Intron | 48590 - 50107 | 38916095-38917612 | 1518 | sequence | - |
| 15 | Exon | 50108 - 51248 | 38914954-38916094 | 1141 | sequence | download |












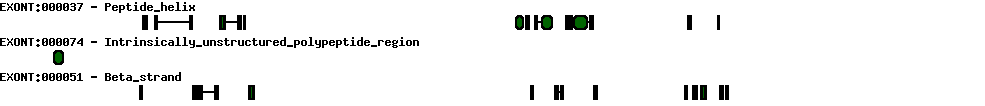



Please wait while loading ...