
| Gene |  |
||
|---|---|---|---|
| Symbol | RBM6 | ||
| Synonyms | 3G2, DEF-3, DEF3, g16, NY-LU-12 | ||
| Description | RNA binding motif protein 6 | ||
| EnsEMBL link | ENSG00000004534 | ||
| Chromosome | 3 - forward strand | ||
| Chromosomal location (UCSC link) |
49977477 - 50156390 (178914bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_001167582 NM_005777 AK291719 AK298108 AF069517 AL832236 BX648535 AK308794 AK303371 AK093836 AF091264 AY216800 BC030057 EF566884 EF566885 AF042857 BC046643 EF566883 U50839 BC012112 NM_005778 AF091263 AK314032 AK302766 AK297445 AK297249 BC002957 AB208813 AF103802 AF107493 AB586690 CR618916 U23946 |
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 51 | 333 | AK297445 |
|
| ASS2 | 6 | 272 | NM_005777 AK291719 AF069517 AK303371 AF091264 AF042857 U50839 BC012112 |
|
| ASS3 | 29 | 101 | AK297445 |
|
| ASS4 | 18 | 96 | BC030057 |
|
| ASS5 | 14 | 17 | NM_001167582 NM_005777 AK291719 AK298108 AF069517 AL832236 BX648535 AK308794 AK093836 AF091264 EF566885 AF042857 BC046643 EF566883 U50839 BC012112 |
|
| ASS6 | 38 | 2 | AB208813 |
|
| Alternative 5' splice sites (donor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 6 | -935 | BC030057 |
|
| ASS2 | 1 | -772 | NM_001167582 NM_005777 AK291719 AK298108 AF069517 AL832236 BX648535 AK308794 AK303371 AK093836 AF091264 BC030057 EF566884 EF566885 AF042857 |
|
| ASS3 | 50 | -83 | AK297445 |
|
| ASS4 | 28 | -11 | EF566884 EF566885 EF566883 NM_005778 AF091263 AK314032 AK297445 AK297249 BC002957 AB208813 AF103802 AF107493 AB586690 CR618916 U23946 |
|
| ASS5 | 25 | -1 | BC012112 |
|
| Intron retentions |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| IR1 | 17 | BX648535 |
| IR2 | 30 | AF107493 AB586690 |
| IR3 | 31 | AB208813 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 49977477 | NM_001167582 AK291719 NM_005777 |
| P2 | 1 | 20 | 49977496 | AK298108 |
| P3 | 1 | 107 | 49977583 | AK308794 |
| P4 | 1 | 108 | 49977584 | AK303371 |
| P5 | 1 | 116 | 49977592 | AK093836 |
| P6 | 26 | 148876 | 50126352 | NM_005778 |
| P7 | 26 | 148911 | 50126387 | AK314032 AK302766 AK297445 AK297249 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_001167582 | sequence |
| NM_005777 | sequence |
| AK291719 | sequence |
| AK298108 | sequence |
| AF069517 | sequence |
| AL832236 | sequence |
| BX648535 | sequence |
| AK308794 | sequence |
| AK303371 | sequence |
| AK093836 | sequence |
| AF091264 | sequence |
| AY216800 | sequence |
| BC030057 | sequence |
| EF566884 | sequence |
| EF566885 | sequence |
| AF042857 | sequence |
| BC046643 | sequence |
| EF566883 | sequence |
| U50839 | sequence |
| BC012112 | sequence |
| NM_005778 | sequence |
| AF091263 | sequence |
| AK314032 | sequence |
| AK302766 | sequence |
| AK297445 | sequence |
| AK297249 | sequence |
| BC002957 | sequence |
| AB208813 | sequence |
| AF103802 | sequence |
| AF107493 | sequence |
| AB586690 | sequence |
| CR618916 | sequence |
| U23946 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 4 | 2817 | 49980293 | yes | AY216800 | Cdna | |
| PA2 | 12 | 117810 | 50095286 | no | AK303371 | Full_length | |
| PA3 | 21 | 126399 | 50103875 | AATAAA | no | AK308794 | Full_length |
| PA4 | 25 | 137209 | 50114685 | AATAAA | no | AF069517 | Cdna |
| PA5 | 25 | 137209 | 50114685 | AATAAA | yes | AF042857 | Cdna |
| PA6 | 31 | 162138 | 50139614 | yes | AF107493 | Cdna | |
| PA7 | 31 | 162138 | 50139614 | yes | AB586690 | Cdna | |
| PA8 | 51 | 178413 | 50155889 | no | AK314032 | Full_length | |
| PA9 | 51 | 178564 | 50156040 | no | AK297249 | Full_length | |
| PA10 | 51 | 178753 | 50156229 | no | AK297445 | Full_length | |
| PA11 | 51 | 178914 | 50156390 | yes | NM_005778 | Cdna | |
| PA12 | 51 | 178914 | 50156390 | yes | AF091263 | Cdna | |
| PA13 | 51 | 178914 | 50156390 | yes | BC002957 | Cdna | |
| PA14 | 51 | 178914 | 50156390 | no | AB208813 | Cdna | |
| PA15 | 51 | 178914 | 50156390 | yes | AF103802 | Cdna | |
| PA16 | 51 | 178914 | 50156390 | no | AK302766 | Full_length | |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 5 |
| 26 | 2 |
| Transcription termination & last exon(s) | |
| 4 | 1 |
| 12 | 1 |
| 21 | 1 |
| 25 | 2 |
| 31 | 2 |
| 51 | 9 |
| Exon skipping | |
| 2 | 15 |
| 3 | 15 |
| 4 | 15 |
| 6 | 9 |
| 7 | 6 |
| 8 | 6 |
| 9 | 10 |
| 10 | 2 |
| 11 | 1 |
| 12 | 1 |
| 13 | 1 |
| 14 | 1 |
| 15 | 1 |
| 16 | 1 |
| 17 | 1 |
| 20 | 17 |
| 24 | 3 |
| 25 | 3 |
| 26 | 3 |
| 29 | 1 |
| 31 | 1 |
| 37 | 10 |
| Alternative 3' splice sites | |
| 6 | 1 |
| 14 | 1 |
| 18 | 1 |
| 29 | 1 |
| 38 | 1 |
| 51 | 1 |
| Alternative 5' splice sites | |
| 1 | 1 |
| 6 | 1 |
| 25 | 1 |
| 28 | 1 |
| 50 | 1 |
| Intron retentions | |
| 17 | 1 |
| 30 | 2 |
| 31 | 1 |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 954 | 49977477 - 49978430 | 954 | sequence | download |
| 1 | Intron | 955 - 1786 | 49978431 - 49979262 | 832 | sequence | - |
| 2 | Exon | 1787 - 1902 | 49979263 - 49979378 | 116 | sequence | download |
| 2 | Intron | 1903 - 1968 | 49979379 - 49979444 | 66 | sequence | - |
| 3 | Exon | 1969 - 1983 | 49979445 - 49979459 | 15 | sequence | download |
| 3 | Intron | 1984 - 2048 | 49979460 - 49979524 | 65 | sequence | - |
| 4 | Exon | 2049 - 2817 | 49979525 - 49980293 | 769 | sequence | download |
| 4 | Intron | 2818 - 22532 | 49980294 - 50000008 | 19715 | sequence | - |
| 5 | Exon | 22533 - 22642 | 50000009 - 50000118 | 110 | sequence | download |
| 5 | Intron | 22643 - 27154 | 50000119 - 50004630 | 4512 | sequence | - |
| 6 | Exon | 27155 - 28705 | 50004631 - 50006181 | 1551 | sequence | download |
| 6 | Intron | 28706 - 32015 | 50006182 - 50009491 | 3310 | sequence | - |
| 7 | Exon | 32016 - 32105 | 50009492 - 50009581 | 90 | sequence | download |
| 7 | Intron | 32106 - 35279 | 50009582 - 50012755 | 3174 | sequence | - |
| 8 | Exon | 35280 - 35349 | 50012756 - 50012825 | 70 | sequence | download |
| 8 | Intron | 35350 - 59396 | 50012826 - 50036872 | 24047 | sequence | - |
| 9 | Exon | 59397 - 59470 | 50036873 - 50036946 | 74 | sequence | download |
| 9 | Intron | 59471 - 108201 | 50036947 - 50085677 | 48731 | sequence | - |
| 10 | Exon | 108202 - 108276 | 50085678 - 50085752 | 75 | sequence | download |
| 10 | Intron | 108277 - 114291 | 50085753 - 50091767 | 6015 | sequence | - |
| 11 | Exon | 114292 - 114352 | 50091768 - 50091828 | 61 | sequence | download |
| 11 | Intron | 114353 - 117684 | 50091829 - 50095160 | 3332 | sequence | - |
| 12 | Exon | 117685 - 117960 | 50095161 - 50095436 | 276 | sequence | download |
| 12 | Intron | 117961 - 118358 | 50095437 - 50095834 | 398 | sequence | - |
| 13 | Exon | 118359 - 118519 | 50095835 - 50095995 | 161 | sequence | download |
| 13 | Intron | 118520 - 119588 | 50095996 - 50097064 | 1069 | sequence | - |
| 14 | Exon | 119589 - 119703 | 50097065 - 50097179 | 115 | sequence | download |
| 14 | Intron | 119704 - 120912 | 50097180 - 50098388 | 1209 | sequence | - |
| 15 | Exon | 120913 - 120955 | 50098389 - 50098431 | 43 | sequence | download |
| 15 | Intron | 120956 - 121096 | 50098432 - 50098572 | 141 | sequence | - |
| 16 | Exon | 121097 - 121178 | 50098573 - 50098654 | 82 | sequence | download |
| 16 | Intron | 121179 - 121418 | 50098655 - 50098894 | 240 | sequence | - |
| 17 | Exon | 121419 - 121504 | 50098895 - 50098980 | 86 | sequence | download |
| 17 | Intron | 121505 - 121918 | 50098981 - 50099394 | 414 | sequence | - |
| 18 | Exon | 121919 - 122065 | 50099395 - 50099541 | 147 | sequence | download |
| 18 | Intron | 122066 - 124987 | 50099542 - 50102463 | 2922 | sequence | - |
| 19 | Exon | 124988 - 125083 | 50102464 - 50102559 | 96 | sequence | download |
| 19 | Intron | 125084 - 125535 | 50102560 - 50103011 | 452 | sequence | - |
| 20 | Exon | 125536 - 125589 | 50103012 - 50103065 | 54 | sequence | download |
| 20 | Intron | 125590 - 126198 | 50103066 - 50103674 | 609 | sequence | - |
| 21 | Exon | 126199 - 126459 | 50103675 - 50103935 | 261 | sequence | download |
| 21 | Intron | 126460 - 128646 | 50103936 - 50106122 | 2187 | sequence | - |
| 22 | Exon | 128647 - 128721 | 50106123 - 50106197 | 75 | sequence | download |
| 22 | Intron | 128722 - 130411 | 50106198 - 50107887 | 1690 | sequence | - |
| 23 | Exon | 130412 - 130509 | 50107888 - 50107985 | 98 | sequence | download |
| 23 | Intron | 130510 - 135157 | 50107986 - 50112633 | 4648 | sequence | - |
| 24 | Exon | 135158 - 135287 | 50112634 - 50112763 | 130 | sequence | download |
| 24 | Intron | 135288 - 136964 | 50112764 - 50114440 | 1677 | sequence | - |
| 25 | Exon | 136965 - 137209 | 50114441 - 50114685 | 245 | sequence | download |
| 25 | Intron | 137210 - 148875 | 50114686 - 50126351 | 11666 | sequence | - |
| 26 | Exon | 148876 - 148986 | 50126352 - 50126462 | 111 | sequence | download |
| 26 | Intron | 148987 - 150338 | 50126463 - 50127814 | 1352 | sequence | - |
| 27 | Exon | 150339 - 150408 | 50127815 - 50127884 | 70 | sequence | download |
| 27 | Intron | 150409 - 151999 | 50127885 - 50129475 | 1591 | sequence | - |
| 28 | Exon | 152000 - 152176 | 50129476 - 50129652 | 177 | sequence | download |
| 28 | Intron | 152177 - 153676 | 50129653 - 50131152 | 1500 | sequence | - |
| 29 | Exon | 153677 - 153832 | 50131153 - 50131308 | 156 | sequence | download |
| 29 | Intron | 153833 - 159938 | 50131309 - 50137414 | 6106 | sequence | - |
| 30 | Exon | 159939 - 160008 | 50137415 - 50137484 | 70 | sequence | download |
| 30 | Intron | 160009 - 160488 | 50137485 - 50137964 | 480 | sequence | - |
| 31 | Exon | 160489 - 160562 | 50137965 - 50138038 | 74 | sequence | download |
| 31 | Intron | 160563 - 163039 | 50138039 - 50140515 | 2477 | sequence | - |
| 32 | Exon | 163040 - 163123 | 50140516 - 50140599 | 84 | sequence | download |
| 32 | Intron | 163124 - 164204 | 50140600 - 50141680 | 1081 | sequence | - |
| 33 | Exon | 164205 - 164265 | 50141681 - 50141741 | 61 | sequence | download |
| 33 | Intron | 164266 - 165033 | 50141742 - 50142509 | 768 | sequence | - |
| 34 | Exon | 165034 - 165099 | 50142510 - 50142575 | 66 | sequence | download |
| 34 | Intron | 165100 - 165505 | 50142576 - 50142981 | 406 | sequence | - |
| 35 | Exon | 165506 - 165666 | 50142982 - 50143142 | 161 | sequence | download |
| 35 | Intron | 165667 - 166723 | 50143143 - 50144199 | 1057 | sequence | - |
| 36 | Exon | 166724 - 166821 | 50144200 - 50144297 | 98 | sequence | download |
| 36 | Intron | 166822 - 166974 | 50144298 - 50144450 | 153 | sequence | - |
| 37 | Exon | 166975 - 167048 | 50144451 - 50144524 | 74 | sequence | download |
| 37 | Intron | 167049 - 167438 | 50144525 - 50144914 | 390 | sequence | - |
| 38 | Exon | 167439 - 167526 | 50144915 - 50145002 | 88 | sequence | download |
| 38 | Intron | 167527 - 168026 | 50145003 - 50145502 | 500 | sequence | - |
| 39 | Exon | 168027 - 168104 | 50145503 - 50145580 | 78 | sequence | download |
| 39 | Intron | 168105 - 168188 | 50145581 - 50145664 | 84 | sequence | - |
| 40 | Exon | 168189 - 168261 | 50145665 - 50145737 | 73 | sequence | download |
| 40 | Intron | 168262 - 169559 | 50145738 - 50147035 | 1298 | sequence | - |
| 41 | Exon | 169560 - 169645 | 50147036 - 50147121 | 86 | sequence | download |
| 41 | Intron | 169646 - 170335 | 50147122 - 50147811 | 690 | sequence | - |
| 42 | Exon | 170336 - 170420 | 50147812 - 50147896 | 85 | sequence | download |
| 42 | Intron | 170421 - 170635 | 50147897 - 50148111 | 215 | sequence | - |
| 43 | Exon | 170636 - 170727 | 50148112 - 50148203 | 92 | sequence | download |
| 43 | Intron | 170728 - 173339 | 50148204 - 50150815 | 2612 | sequence | - |
| 44 | Exon | 173340 - 173501 | 50150816 - 50150977 | 162 | sequence | download |
| 44 | Intron | 173502 - 173906 | 50150978 - 50151382 | 405 | sequence | - |
| 45 | Exon | 173907 - 174056 | 50151383 - 50151532 | 150 | sequence | download |
| 45 | Intron | 174057 - 174136 | 50151533 - 50151612 | 80 | sequence | - |
| 46 | Exon | 174137 - 174208 | 50151613 - 50151684 | 72 | sequence | download |
| 46 | Intron | 174209 - 175384 | 50151685 - 50152860 | 1176 | sequence | - |
| 47 | Exon | 175385 - 175564 | 50152861 - 50153040 | 180 | sequence | download |
| 47 | Intron | 175565 - 175862 | 50153041 - 50153338 | 298 | sequence | - |
| 48 | Exon | 175863 - 175937 | 50153339 - 50153413 | 75 | sequence | download |
| 48 | Intron | 175938 - 177030 | 50153414 - 50154506 | 1093 | sequence | - |
| 49 | Exon | 177031 - 177128 | 50154507 - 50154604 | 98 | sequence | download |
| 49 | Intron | 177129 - 177206 | 50154605 - 50154682 | 78 | sequence | - |
| 50 | Exon | 177207 - 177336 | 50154683 - 50154812 | 130 | sequence | download |
| 50 | Intron | 177337 - 178287 | 50154813 - 50155763 | 951 | sequence | - |
| 51 | Exon | 178288 - 178914 | 50155764 - 50156390 | 627 | sequence | download |

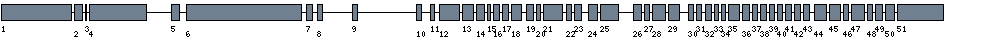




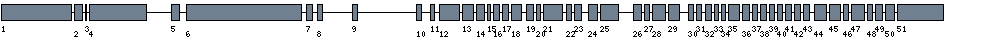
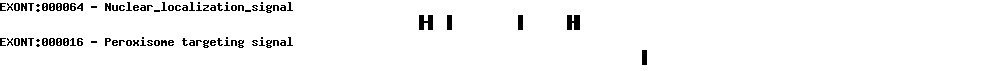
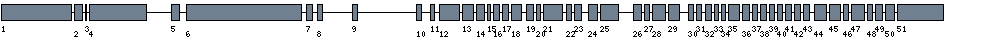

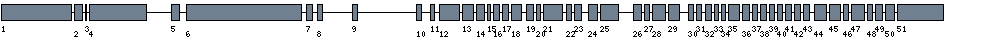

Please wait while loading ...