
 FasterDB identifier: 14439 - SEMA3F (Sema4, SEMAK)
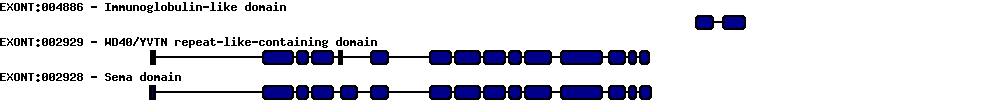
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
FasterDB identifier: 14439 - SEMA3F (Sema4, SEMAK)
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F

| Gene |  |
||
|---|---|---|---|
| Symbol | SEMA3F | ||
| Synonyms | Sema4, SEMAK | ||
| Description | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F | ||
| EnsEMBL link | ENSG00000001617 | ||
| Chromosome | 3 - forward strand | ||
| Chromosomal location (UCSC link) |
50192524 - 50226507 (33984bp) | ||
| Sequence | sequence | ||
| Associated transcripts | AB209259 BC042914 NM_004186 U33920 U38276 |
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 5 | 12 | BC042914 NM_004186 U33920 U38276 |
|
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 50192524 | AB209259 |
| P2 | 2 | 325 | 50192848 | BC042914 NM_004186 |
| P3 | 3 | 3929 | 50196452 | U38276 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| AB209259 | sequence |
| BC042914 | sequence |
| NM_004186 | sequence |
| U33920 | sequence |
| U38276 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 21 | 33131 | 50225654 | TATAAA | no | AI378926 | 3prime_est |
| PA2 | 21 | 33984 | 50226507 | AATAAA | no | AB209259 | Cdna |
| PA3 | 21 | 33984 | 50226507 | AATAAA | yes | BC042914 | Cdna |
| PA4 | 21 | 33984 | 50226507 | AATAAA | yes | NM_004186 | Cdna |
| Exon skipping |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ES1 | 2 | AB209259 |
| ES2 | 3 | AB209259 BC042914 NM_004186 U33920 |
| ES3 | 4 | AB209259 |
| ES4 | 8 | AB209259 U38276 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 1 |
| 2 | 1 |
| 3 | 1 |
| Transcription termination & last exon(s) | |
| 21 | 4 |
| Exon skipping | |
| 2 | 1 |
| 3 | 4 |
| 4 | 1 |
| 8 | 2 |
| Alternative 3' splice sites | |
| 5 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| 16 | Expression | Expression | Expression |
| 17 | Expression | Expression | Expression |
| 18 | Expression | Expression | Expression |
| 19 | Expression | Expression | Expression |
| 20 | Expression | Expression | Expression |
| 21 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 58 | 50192524 - 50192581 | 58 | sequence | download |
| 1 | Intron | 59 - 324 | 50192582 - 50192847 | 266 | sequence | - |
| 2 | Exon | 325 - 474 | 50192848 - 50192997 | 150 | sequence | download |
| 2 | Intron | 475 - 3928 | 50192998 - 50196451 | 3454 | sequence | - |
| 3 | Exon | 3929 - 4198 | 50196452 - 50196721 | 270 | sequence | download |
| 3 | Intron | 4199 - 4484 | 50196722 - 50197007 | 286 | sequence | - |
| 4 | Exon | 4485 - 4644 | 50197008 - 50197167 | 160 | sequence | download |
| 4 | Intron | 4645 - 18690 | 50197168 - 50211213 | 14046 | sequence | - |
| 5 | Exon | 18691 - 18863 | 50211214 - 50211386 | 173 | sequence | download |
| 5 | Intron | 18864 - 18961 | 50211387 - 50211484 | 98 | sequence | - |
| 6 | Exon | 18962 - 19024 | 50211485 - 50211547 | 63 | sequence | download |
| 6 | Intron | 19025 - 19140 | 50211548 - 50211663 | 116 | sequence | - |
| 7 | Exon | 19141 - 19260 | 50211664 - 50211783 | 120 | sequence | download |
| 7 | Intron | 19261 - 20005 | 50211784 - 50212528 | 745 | sequence | - |
| 8 | Exon | 20006 - 20098 | 50212529 - 50212621 | 93 | sequence | download |
| 8 | Intron | 20099 - 21677 | 50212622 - 50214200 | 1579 | sequence | - |
| 9 | Exon | 21678 - 21771 | 50214201 - 50214294 | 94 | sequence | download |
| 9 | Intron | 21772 - 27193 | 50214295 - 50219716 | 5422 | sequence | - |
| 10 | Exon | 27194 - 27313 | 50219717 - 50219836 | 120 | sequence | download |
| 10 | Intron | 27314 - 27553 | 50219837 - 50220076 | 240 | sequence | - |
| 11 | Exon | 27554 - 27693 | 50220077 - 50220216 | 140 | sequence | download |
| 11 | Intron | 27694 - 27813 | 50220217 - 50220336 | 120 | sequence | - |
| 12 | Exon | 27814 - 27928 | 50220337 - 50220451 | 115 | sequence | download |
| 12 | Intron | 27929 - 28095 | 50220452 - 50220618 | 167 | sequence | - |
| 13 | Exon | 28096 - 28165 | 50220619 - 50220688 | 70 | sequence | download |
| 13 | Intron | 28166 - 28329 | 50220689 - 50220852 | 164 | sequence | - |
| 14 | Exon | 28330 - 28474 | 50220853 - 50220997 | 145 | sequence | download |
| 14 | Intron | 28475 - 29501 | 50220998 - 50222024 | 1027 | sequence | - |
| 15 | Exon | 29502 - 29724 | 50222025 - 50222247 | 223 | sequence | download |
| 15 | Intron | 29725 - 30352 | 50222248 - 50222875 | 628 | sequence | - |
| 16 | Exon | 30353 - 30441 | 50222876 - 50222964 | 89 | sequence | download |
| 16 | Intron | 30442 - 30575 | 50222965 - 50223098 | 134 | sequence | - |
| 17 | Exon | 30576 - 30617 | 50223099 - 50223140 | 42 | sequence | download |
| 17 | Intron | 30618 - 30798 | 50223141 - 50223321 | 181 | sequence | - |
| 18 | Exon | 30799 - 30956 | 50223322 - 50223479 | 158 | sequence | download |
| 18 | Intron | 30957 - 31190 | 50223480 - 50223713 | 234 | sequence | - |
| 19 | Exon | 31191 - 31258 | 50223714 - 50223781 | 68 | sequence | download |
| 19 | Intron | 31259 - 31522 | 50223782 - 50224045 | 264 | sequence | - |
| 20 | Exon | 31523 - 31656 | 50224046 - 50224179 | 134 | sequence | download |
| 20 | Intron | 31657 - 32614 | 50224180 - 50225137 | 958 | sequence | - |
| 21 | Exon | 32615 - 33984 | 50225138 - 50226507 | 1370 | sequence | download |

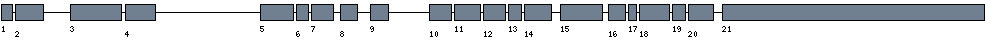



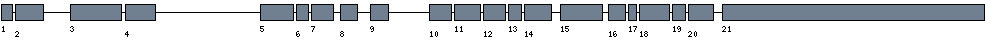

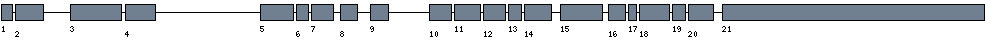

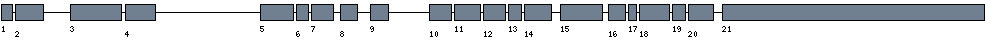
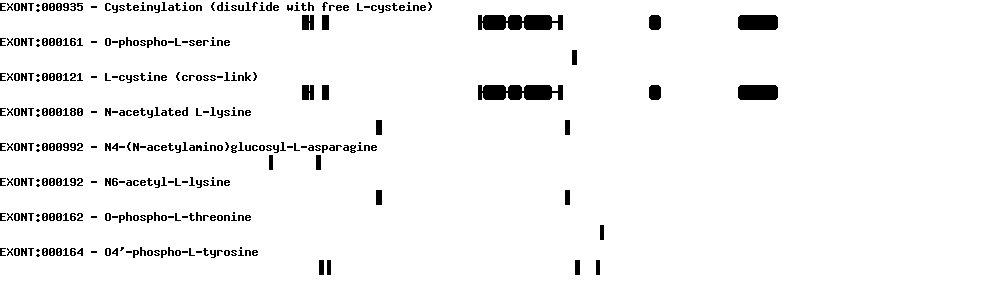
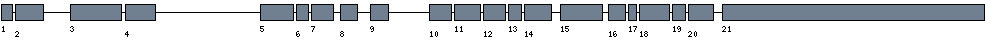

Please wait while loading ...