
| Gene |  |
||
|---|---|---|---|
| Symbol | TRAIP | ||
| Synonyms | RNF206, TRIP | ||
| Description | TRAF interacting protein | ||
| EnsEMBL link | ENSG00000183763 | ||
| Chromosome | 3 - reverse strand | ||
| Chromosomal location (UCSC link) |
49866028 - 49893997 (27970bp) | ||
| Sequence | sequence | ||
| Associated transcripts | AK295780 U77845 NM_005879 AK292172 BC019283 BC000310 AK225873 CR623917 AB451320 AB451449 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 49893997 | AK295780 |
| P2 | 1 | 18 | 49893980 | AK292172 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| AK295780 | sequence |
| U77845 | sequence |
| NM_005879 | sequence |
| AK292172 | sequence |
| BC019283 | sequence |
| BC000310 | sequence |
| AK225873 | sequence |
| CR623917 | sequence |
| AB451320 | sequence |
| AB451449 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 5 | 13420 | 49880578 | no | AK295780 | Full_length | |
| PA2 | 15 | 27818 | 49866180 | no | AK292172 | Full_length | |
| PA3 | 15 | 27970 | 49866028 | ATTAAA | yes | NM_005879 | Cdna |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 2 |
| Transcription termination & last exon(s) | |
| 5 | 1 |
| 15 | 2 |
| Exon skipping | |
| 6 | 1 |
| 7 | 1 |
| 8 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 209 | 49893789-49893997 | 209 | sequence | download |
| 1 | Intron | 210 - 8364 | 49885634-49893788 | 8155 | sequence | - |
| 2 | Exon | 8365 - 8422 | 49885576-49885633 | 58 | sequence | download |
| 2 | Intron | 8423 - 8956 | 49885042-49885575 | 534 | sequence | - |
| 3 | Exon | 8957 - 9040 | 49884958-49885041 | 84 | sequence | download |
| 3 | Intron | 9041 - 11984 | 49882014-49884957 | 2944 | sequence | - |
| 4 | Exon | 11985 - 12024 | 49881974-49882013 | 40 | sequence | download |
| 4 | Intron | 12025 - 12636 | 49881362-49881973 | 612 | sequence | - |
| 5 | Exon | 12637 - 12764 | 49881234-49881361 | 128 | sequence | download |
| 5 | Intron | 12765 - 14017 | 49879981-49881233 | 1253 | sequence | - |
| 6 | Exon | 14018 - 14112 | 49879886-49879980 | 95 | sequence | download |
| 6 | Intron | 14113 - 14625 | 49879373-49879885 | 513 | sequence | - |
| 7 | Exon | 14626 - 14739 | 49879259-49879372 | 114 | sequence | download |
| 7 | Intron | 14740 - 15492 | 49878506-49879258 | 753 | sequence | - |
| 8 | Exon | 15493 - 15580 | 49878418-49878505 | 88 | sequence | download |
| 8 | Intron | 15581 - 16191 | 49877807-49878417 | 611 | sequence | - |
| 9 | Exon | 16192 - 16281 | 49877717-49877806 | 90 | sequence | download |
| 9 | Intron | 16282 - 16704 | 49877294-49877716 | 423 | sequence | - |
| 10 | Exon | 16705 - 16793 | 49877205-49877293 | 89 | sequence | download |
| 10 | Intron | 16794 - 24496 | 49869502-49877204 | 7703 | sequence | - |
| 11 | Exon | 24497 - 24649 | 49869349-49869501 | 153 | sequence | download |
| 11 | Intron | 24650 - 26496 | 49867502-49869348 | 1847 | sequence | - |
| 12 | Exon | 26497 - 26545 | 49867453-49867501 | 49 | sequence | download |
| 12 | Intron | 26546 - 26798 | 49867200-49867452 | 253 | sequence | - |
| 13 | Exon | 26799 - 26948 | 49867050-49867199 | 150 | sequence | download |
| 13 | Intron | 26949 - 27056 | 49866942-49867049 | 108 | sequence | - |
| 14 | Exon | 27057 - 27107 | 49866891-49866941 | 51 | sequence | download |
| 14 | Intron | 27108 - 27339 | 49866659-49866890 | 232 | sequence | - |
| 15 | Exon | 27340 - 27970 | 49866028-49866658 | 631 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-328 | 12936 - 12958 | 49881040 - 49881062 | miranda |
| M2 | hsa-miR-149 | 13138 - 13160 | 49880838 - 49880860 | miranda |
| M3 | hsa-miR-708 | 13151 - 13175 | 49880823 - 49880847 | miranda |
| Group 2 | ||||
| M1 | hsa-miR-370 | 27687 - 27694 | 49866304 - 49866311 | targetscan |
| Group 3 | ||||
| M1 | hsa-miR-590-3p | 27929 - 27950 | 49866048 - 49866069 | miranda |
| M2 | hsa-miR-590-3p | 27942 - 27949 | 49866049 - 49866056 | pita |














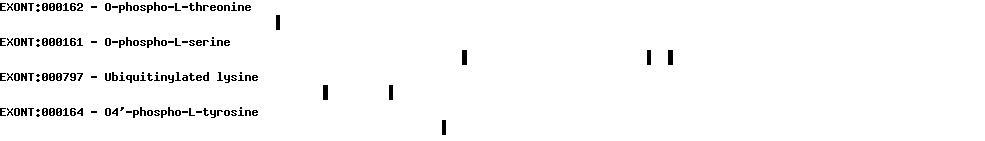


Please wait while loading ...