
 FasterDB identifier: 13185 - UBA7 (D8, UBA1B, UBE1L, UBE2)
ubiquitin-like modifier activating enzyme 7
FasterDB identifier: 13185 - UBA7 (D8, UBA1B, UBE1L, UBE2)
ubiquitin-like modifier activating enzyme 7

| Gene |  |
||
|---|---|---|---|
| Symbol | UBA7 | ||
| Synonyms | D8, UBA1B, UBE1L, UBE2 | ||
| Description | ubiquitin-like modifier activating enzyme 7 | ||
| EnsEMBL link | ENSG00000182179 | ||
| Chromosome | 3 - reverse strand | ||
| Chromosomal location (UCSC link) |
49842639 - 49851391 (8753bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_003335 L13852 BC006378 BT007026 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 49851391 | NM_003335 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_003335 | sequence |
| L13852 | sequence |
| BC006378 | sequence |
| BT007026 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 24 | 8753 | 49842639 | AATAAA | yes | NM_003335 | Cdna |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 1 |
| Transcription termination & last exon(s) | |
| 24 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| 16 | Expression | Expression | Expression |
| 17 | Expression | Expression | Expression |
| 18 | Expression | Expression | Expression |
| 19 | Expression | Expression | Expression |
| 20 | Expression | Expression | Expression |
| 21 | Expression | Expression | Expression |
| 22 | Expression | Expression | Expression |
| 23 | Expression | Expression | Expression |
| 24 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 227 | 49851165-49851391 | 227 | sequence | download |
| 1 | Intron | 228 - 311 | 49851081-49851164 | 84 | sequence | - |
| 2 | Exon | 312 - 480 | 49850912-49851080 | 169 | sequence | download |
| 2 | Intron | 481 - 575 | 49850817-49850911 | 95 | sequence | - |
| 3 | Exon | 576 - 710 | 49850682-49850816 | 135 | sequence | download |
| 3 | Intron | 711 - 790 | 49850602-49850681 | 80 | sequence | - |
| 4 | Exon | 791 - 897 | 49850495-49850601 | 107 | sequence | download |
| 4 | Intron | 898 - 1220 | 49850172-49850494 | 323 | sequence | - |
| 5 | Exon | 1221 - 1311 | 49850081-49850171 | 91 | sequence | download |
| 5 | Intron | 1312 - 1415 | 49849977-49850080 | 104 | sequence | - |
| 6 | Exon | 1416 - 1551 | 49849841-49849976 | 136 | sequence | download |
| 6 | Intron | 1552 - 1752 | 49849640-49849840 | 201 | sequence | - |
| 7 | Exon | 1753 - 1850 | 49849542-49849639 | 98 | sequence | download |
| 7 | Intron | 1851 - 1942 | 49849450-49849541 | 92 | sequence | - |
| 8 | Exon | 1943 - 2089 | 49849303-49849449 | 147 | sequence | download |
| 8 | Intron | 2090 - 2503 | 49848889-49849302 | 414 | sequence | - |
| 9 | Exon | 2504 - 2686 | 49848706-49848888 | 183 | sequence | download |
| 9 | Intron | 2687 - 2867 | 49848525-49848705 | 181 | sequence | - |
| 10 | Exon | 2868 - 2975 | 49848417-49848524 | 108 | sequence | download |
| 10 | Intron | 2976 - 3126 | 49848266-49848416 | 151 | sequence | - |
| 11 | Exon | 3127 - 3207 | 49848185-49848265 | 81 | sequence | download |
| 11 | Intron | 3208 - 3286 | 49848106-49848184 | 79 | sequence | - |
| 12 | Exon | 3287 - 3442 | 49847950-49848105 | 156 | sequence | download |
| 12 | Intron | 3443 - 3530 | 49847862-49847949 | 88 | sequence | - |
| 13 | Exon | 3531 - 3696 | 49847696-49847861 | 166 | sequence | download |
| 13 | Intron | 3697 - 3775 | 49847617-49847695 | 79 | sequence | - |
| 14 | Exon | 3776 - 3981 | 49847411-49847616 | 206 | sequence | download |
| 14 | Intron | 3982 - 4079 | 49847313-49847410 | 98 | sequence | - |
| 15 | Exon | 4080 - 4144 | 49847248-49847312 | 65 | sequence | download |
| 15 | Intron | 4145 - 4233 | 49847159-49847247 | 89 | sequence | - |
| 16 | Exon | 4234 - 4417 | 49846975-49847158 | 184 | sequence | download |
| 16 | Intron | 4418 - 4494 | 49846898-49846974 | 77 | sequence | - |
| 17 | Exon | 4495 - 4569 | 49846823-49846897 | 75 | sequence | download |
| 17 | Intron | 4570 - 4799 | 49846593-49846822 | 230 | sequence | - |
| 18 | Exon | 4800 - 4983 | 49846409-49846592 | 184 | sequence | download |
| 18 | Intron | 4984 - 5490 | 49845902-49846408 | 507 | sequence | - |
| 19 | Exon | 5491 - 5573 | 49845819-49845901 | 83 | sequence | download |
| 19 | Intron | 5574 - 5846 | 49845546-49845818 | 273 | sequence | - |
| 20 | Exon | 5847 - 5939 | 49845453-49845545 | 93 | sequence | download |
| 20 | Intron | 5940 - 6031 | 49845361-49845452 | 92 | sequence | - |
| 21 | Exon | 6032 - 6223 | 49845169-49845360 | 192 | sequence | download |
| 21 | Intron | 6224 - 7793 | 49843599-49845168 | 1570 | sequence | - |
| 22 | Exon | 7794 - 7886 | 49843506-49843598 | 93 | sequence | download |
| 22 | Intron | 7887 - 7961 | 49843431-49843505 | 75 | sequence | - |
| 23 | Exon | 7962 - 8062 | 49843330-49843430 | 101 | sequence | download |
| 23 | Intron | 8063 - 8521 | 49842871-49843329 | 459 | sequence | - |
| 24 | Exon | 8522 - 8753 | 49842639-49842870 | 232 | sequence | download |


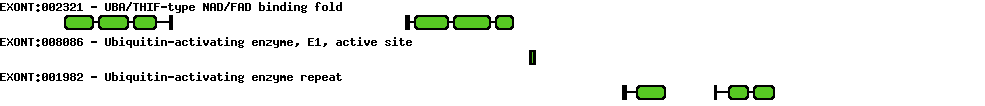









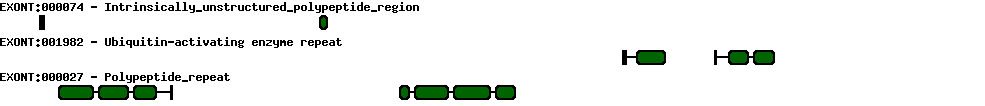

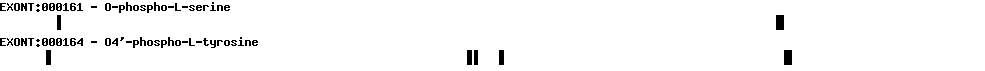

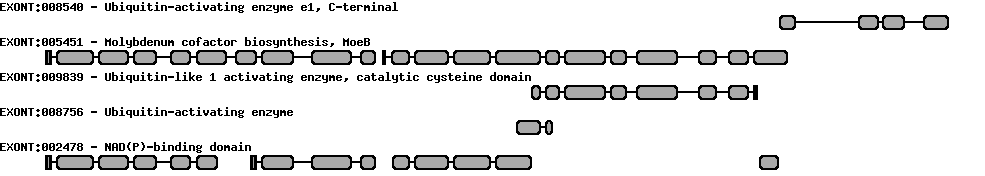
Please wait while loading ...