
| Gene |  |
||
|---|---|---|---|
| Symbol | CAMKV | ||
| Synonyms | MGC8407, VACAMKL | ||
| Description | CaM kinase-like vesicle-associated | ||
| EnsEMBL link | ENSG00000164076 | ||
| Chromosome | 3 - reverse strand | ||
| Chromosomal location (UCSC link) |
49895421 - 49907388 (11968bp) | ||
| Sequence | sequence | ||
| Associated transcripts | AK315525 AK074856 NM_024046 AK075294 AK295588 AK074899 AK297258 AK299953 AL136697 BX647074 AK297437 BC017363 BC001921 BC005828 BC000497 BC019256 CR533508 AK226073 |
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 5 | 40 | AK299953 |
|
| ASS2 | 8 | 8 | AL136697 CR533508 |
|
| ASS3 | 6 | 7 | AK315525 AK074856 NM_024046 AK075294 AK074899 AK297258 AK299953 AL136697 BX647074 AK297437 BC017363 BC001921 BC005828 BC000497 BC019256 CR533508 |
|
| Alternative 5' splice sites (donor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 3 | -41 | AK297437 |
|
| ASS2 | 2 | -38 | AK297258 |
|
| ASS3 | 4 | -4 | AK315525 AK074856 NM_024046 AK075294 AK074899 AK297258 AK299953 AL136697 BX647074 AK297437 BC017363 BC001921 BC005828 BC000497 BC019256 CR533508 |
|
| Intron retentions |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| IR1 | 6 | BX647074 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 49907388 | AK315525 |
| P2 | 1 | 20 | 49907369 | AK074856 |
| P3 | 1 | 39 | 49907350 | AK075294 |
| P4 | 1 | 47 | 49907342 | AK295588 |
| P5 | 1 | 50 | 49907339 | AK074899 AK297258 |
| P6 | 1 | 51 | 49907338 | AK299953 |
| P7 | 1 | 53 | 49907336 | AK297437 |
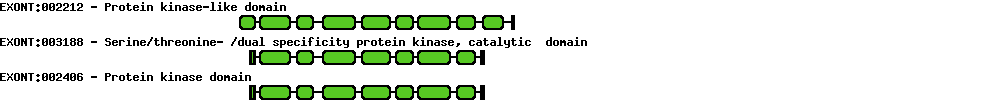
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| AK315525 | sequence |
| AK074856 | sequence |
| NM_024046 | sequence |
| AK075294 | sequence |
| AK295588 | sequence |
| AK074899 | sequence |
| AK297258 | sequence |
| AK299953 | sequence |
| AL136697 | sequence |
| BX647074 | sequence |
| AK297437 | sequence |
| BC017363 | sequence |
| BC001921 | sequence |
| BC005828 | sequence |
| BC000497 | sequence |
| BC019256 | sequence |
| CR533508 | sequence |
| AK226073 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 10 | 9756 | 49897633 | no | AK299953 | Full_length | |
| PA2 | 11 | 10695 | 49896694 | no | AK297258 | Full_length | |
| PA3 | 11 | 10695 | 49896694 | no | AK297437 | Full_length | |
| PA4 | 11 | 11968 | 49895421 | AATAAA | yes | BX647074 | Cdna |
| PA5 | 11 | 12741 | 49894648 | AATAAA | no | DB367081 | 3prime_est |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 8383 | 1 | 1 |
| 8383 | 2 | 2 |
| 8383 | 3 | 3 |
| 8383 | 4 | 4 |
| 8383 | 5 | 5 |
| 8383 | 6 | 6 |
| 8383 | 7 | 7 |
| 8383 | 8 | 8 |
| 8383 | 9 | 9 |
| 8383 | 10 | 10 |
| 8383 | 11 | 11 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 7 |
| Transcription termination & last exon(s) | |
| 10 | 1 |
| 11 | 4 |
| Exon skipping | |
| 5 | 1 |
| 7 | 2 |
| Alternative 3' splice sites | |
| 5 | 1 |
| 6 | 1 |
| 8 | 1 |
| Alternative 5' splice sites | |
| 2 | 1 |
| 3 | 1 |
| 4 | 1 |
| Exon deletions | |
| 11 | 4 |
| Intron retentions | |
| 6 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 198 | 49907191-49907388 | 198 | sequence | download |
| 1 | Intron | 199 - 7553 | 49899836-49907190 | 7355 | sequence | - |
| 2 | Exon | 7554 - 7662 | 49899727-49899835 | 109 | sequence | download |
| 2 | Intron | 7663 - 7779 | 49899610-49899726 | 117 | sequence | - |
| 3 | Exon | 7780 - 7911 | 49899478-49899609 | 132 | sequence | download |
| 3 | Intron | 7912 - 8090 | 49899299-49899477 | 179 | sequence | - |
| 4 | Exon | 8091 - 8169 | 49899220-49899298 | 79 | sequence | download |
| 4 | Intron | 8170 - 8378 | 49899011-49899219 | 209 | sequence | - |
| 5 | Exon | 8379 - 8517 | 49898872-49899010 | 139 | sequence | download |
| 5 | Intron | 8518 - 8648 | 49898741-49898871 | 131 | sequence | - |
| 6 | Exon | 8649 - 8776 | 49898613-49898740 | 128 | sequence | download |
| 6 | Intron | 8777 - 8937 | 49898452-49898612 | 161 | sequence | - |
| 7 | Exon | 8938 - 9013 | 49898376-49898451 | 76 | sequence | download |
| 7 | Intron | 9014 - 9103 | 49898286-49898375 | 90 | sequence | - |
| 8 | Exon | 9104 - 9240 | 49898149-49898285 | 137 | sequence | download |
| 8 | Intron | 9241 - 9406 | 49897983-49898148 | 166 | sequence | - |
| 9 | Exon | 9407 - 9485 | 49897904-49897982 | 79 | sequence | download |
| 9 | Intron | 9486 - 9697 | 49897692-49897903 | 212 | sequence | - |
| 10 | Exon | 9698 - 9785 | 49897604-49897691 | 88 | sequence | download |
| 10 | Intron | 9786 - 10074 | 49897315-49897603 | 289 | sequence | - |
| 11 | Exon | 10075 - 11968 | 49895421-49897314 | 1894 | sequence | download |
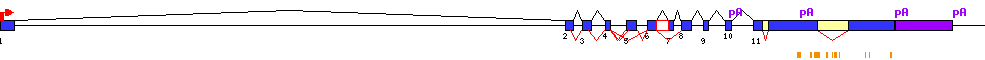
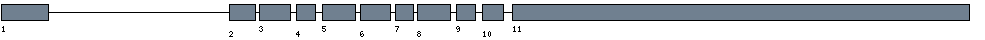

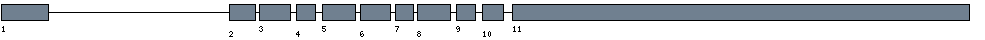


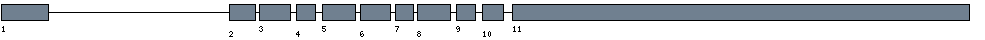
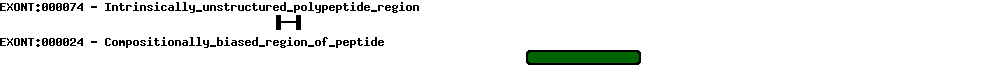
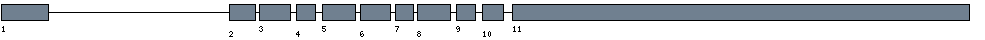

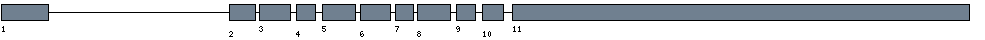

Please wait while loading ...