
| Gene |  |
||
|---|---|---|---|
| Symbol | OPTN | ||
| Synonyms | FIP2, GLC1E, HIP7, HYPL, NRP, TFIIIA-INTP | ||
| Description | optineurin | ||
| EnsEMBL link | ENSG00000123240 | ||
| Chromosome | 10 - forward strand | ||
| Chromosomal location (UCSC link) |
13141449 - 13180276 (38828bp) | ||
| Sequence | sequence | ||
| Associated transcripts | AK055403 NM_001008211 NM_001008213 NM_021980 NM_001008212 AK316171 AK303715 BC032762 BC013876 CR597026 CR603260 CR601600 AF061034 AF420371 AF420372 AF420373 AK122746 AF049614 |
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 6 | 94 | AK055403 NM_001008211 NM_001008213 NM_021980 NM_001008212 BC032762 BC013876 CR597026 CR603260 CR601600 AF061034 AF420371 AF420372 AF420373 |
|
| ASS2 | 9 | 18 | BC032762 CR603260 |
|
| ASS3 | 5 | 15 | AK055403 NM_001008213 NM_021980 NM_001008212 AK316171 AK303715 BC032762 BC013876 CR597026 CR603260 CR601600 AF420372 AF420373 |
|
| Alternative 5' splice sites (donor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 4 | -4 | NM_001008211 NM_001008213 BC032762 AF420371 |
|
| Intron retentions |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| IR1 | 12 | AK122746 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 13141449 | AK055403 |
| P2 | 2 | 634 | 13142082 | NM_001008211 NM_001008213 NM_021980 NM_001008212 |
| P3 | 2 | 688 | 13142136 | AK316171 |
| P4 | 2 | 710 | 13142158 | AK303715 |
| P5 | 11 | 23918 | 13165366 | AK122746 |
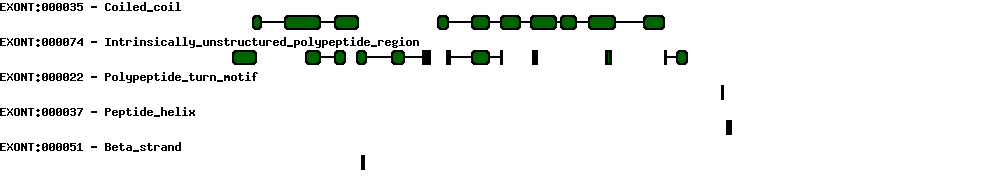
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| AK055403 | sequence |
| NM_001008211 | sequence |
| NM_001008213 | sequence |
| NM_021980 | sequence |
| NM_001008212 | sequence |
| AK316171 | sequence |
| AK303715 | sequence |
| BC032762 | sequence |
| BC013876 | sequence |
| CR597026 | sequence |
| CR603260 | sequence |
| CR601600 | sequence |
| AF061034 | sequence |
| AF420371 | sequence |
| AF420372 | sequence |
| AF420373 | sequence |
| AK122746 | sequence |
| AF049614 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 17 | 37475 | 13178923 | no | AK055403 | Full_length | |
| PA2 | 17 | 37676 | 13179124 | no | AK316171 | Full_length | |
| PA3 | 17 | 37676 | 13179124 | no | AK303715 | Full_length | |
| PA4 | 17 | 38828 | 13180276 | no | NM_001008211 | Cdna | |
| PA5 | 17 | 38828 | 13180276 | no | NM_001008213 | Cdna | |
| PA6 | 17 | 38828 | 13180276 | no | NM_021980 | Cdna | |
| PA7 | 17 | 38828 | 13180276 | no | NM_001008212 | Cdna | |
| PA8 | 17 | 40857 | 13182305 | AATAAA | yes | AI635684 | 3prime_est |
| Exon skipping |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ES1 | 2 | AK055403 |
| ES2 | 3 | NM_021980 AK316171 AK303715 BC013876 CR597026 CR603260 CR601600 AF420372 |
| ES3 | 4 | AK055403 NM_021980 NM_001008212 AK316171 AK303715 BC013876 CR597026 CR603260 CR601600 AF420372 AF420373 |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 9602 | 5 | 2 |
| 9602 | 6 | 3 |
| 9602 | 7 | 4 |
| 9602 | 9 | 6 |
| 9602 | 10 | 8 |
| 9602 | 11 | 9 |
| 9602 | 12 | 10 |
| 9602 | 13 | 11 |
| 9602 | 14 | 12 |
| 9602 | 15 | 13 |
| 9602 | 16 | 14 |
| 9602 | 17 | 15 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 1 |
| 2 | 3 |
| 11 | 1 |
| Transcription termination & last exon(s) | |
| 17 | 8 |
| Exon skipping | |
| 2 | 1 |
| 3 | 8 |
| 4 | 11 |
| Alternative 3' splice sites | |
| 5 | 1 |
| 6 | 1 |
| 9 | 1 |
| Alternative 5' splice sites | |
| 4 | 1 |
| Intron retentions | |
| 12 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| 16 | Expression | Expression | Expression |
| 17 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 510 | 13141449 - 13141958 | 510 | sequence | download |
| 1 | Intron | 511 - 633 | 13141959 - 13142081 | 123 | sequence | - |
| 2 | Exon | 634 - 854 | 13142082 - 13142302 | 221 | sequence | download |
| 2 | Intron | 855 - 8689 | 13142303 - 13150137 | 7835 | sequence | - |
| 3 | Exon | 8690 - 8841 | 13150138 - 13150289 | 152 | sequence | download |
| 3 | Intron | 8842 - 9513 | 13150290 - 13150961 | 672 | sequence | - |
| 4 | Exon | 9514 - 9571 | 13150962 - 13151019 | 58 | sequence | download |
| 4 | Intron | 9572 - 9648 | 13151020 - 13151096 | 77 | sequence | - |
| 5 | Exon | 9649 - 9840 | 13151097 - 13151288 | 192 | sequence | download |
| 5 | Intron | 9841 - 10731 | 13151289 - 13152179 | 891 | sequence | - |
| 6 | Exon | 10732 - 11028 | 13152180 - 13152476 | 297 | sequence | download |
| 6 | Intron | 11029 - 13004 | 13152477 - 13154452 | 1976 | sequence | - |
| 7 | Exon | 13005 - 13187 | 13154453 - 13154635 | 183 | sequence | download |
| 7 | Intron | 13188 - 16818 | 13154636 - 13158266 | 3631 | sequence | - |
| 8 | Exon | 16819 - 16892 | 13158267 - 13158340 | 74 | sequence | download |
| 8 | Intron | 16893 - 19439 | 13158341 - 13160887 | 2547 | sequence | - |
| 9 | Exon | 19440 - 19592 | 13160888 - 13161040 | 153 | sequence | download |
| 9 | Intron | 19593 - 22936 | 13161041 - 13164384 | 3344 | sequence | - |
| 10 | Exon | 22937 - 23039 | 13164385 - 13164487 | 103 | sequence | download |
| 10 | Intron | 23040 - 24546 | 13164488 - 13165994 | 1507 | sequence | - |
| 11 | Exon | 24547 - 24662 | 13165995 - 13166110 | 116 | sequence | download |
| 11 | Intron | 24663 - 25969 | 13166111 - 13167417 | 1307 | sequence | - |
| 12 | Exon | 25970 - 26119 | 13167418 - 13167567 | 150 | sequence | download |
| 12 | Intron | 26120 - 26497 | 13167568 - 13167945 | 378 | sequence | - |
| 13 | Exon | 26498 - 26591 | 13167946 - 13168039 | 94 | sequence | download |
| 13 | Intron | 26592 - 28296 | 13168040 - 13169744 | 1705 | sequence | - |
| 14 | Exon | 28297 - 28455 | 13169745 - 13169903 | 159 | sequence | download |
| 14 | Intron | 28456 - 32618 | 13169904 - 13174066 | 4163 | sequence | - |
| 15 | Exon | 32619 - 32749 | 13174067 - 13174197 | 131 | sequence | download |
| 15 | Intron | 32750 - 34053 | 13174198 - 13175501 | 1304 | sequence | - |
| 16 | Exon | 34054 - 34133 | 13175502 - 13175581 | 80 | sequence | download |
| 16 | Intron | 34134 - 37296 | 13175582 - 13178744 | 3163 | sequence | - |
| 17 | Exon | 37297 - 38828 | 13178745 - 13180276 | 1532 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-432 | 37469 - 37475 | 13178917 - 13178923 | pita |
| Group 2 | ||||
| M1 | hsa-miR-570 | 37496 - 37502 | 13178944 - 13178950 | pita |
| Group 3 | ||||
| M1 | hsa-miR-361-5p | 37944 - 37965 | 13179392 - 13179413 | miranda |
| M2 | hsa-miR-324-5p | 38089 - 38110 | 13179537 - 13179558 | miranda |
| M3 | hsa-miR-9 | 38235 - 38257 | 13179683 - 13179705 | miranda |
| M4 | hsa-miR-9 | 38250 - 38257 | 13179698 - 13179705 | targetscan |
| M5 | hsa-miR-449a | 38432 - 38455 | 13179880 - 13179903 | miranda |
| M6 | hsa-miR-449b | 38432 - 38455 | 13179880 - 13179903 | miranda |
| M7 | hsa-miR-590-3p | 38500 - 38520 | 13179948 - 13179968 | miranda |
| M8 | hsa-miR-590-3p | 38512 - 38519 | 13179960 - 13179967 | pita |
| M9 | hsa-miR-182 | 38738 - 38761 | 13180186 - 13180209 | miranda |
| M10 | hsa-miR-199a-5p | 38755 - 38777 | 13180203 - 13180225 | miranda |
| M11 | hsa-miR-199b-5p | 38755 - 38777 | 13180203 - 13180225 | miranda |













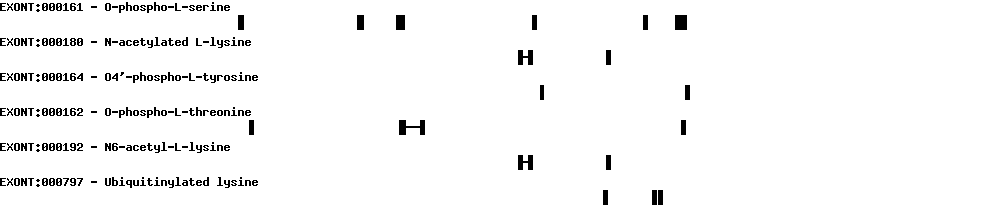


Please wait while loading ...