
 FasterDB identifier: 11765 - MCM10 (CNA43, DNA43, PRO2249)
minichromosome maintenance complex component 10
FasterDB identifier: 11765 - MCM10 (CNA43, DNA43, PRO2249)
minichromosome maintenance complex component 10

| Gene |  |
||
|---|---|---|---|
| Symbol | MCM10 | ||
| Synonyms | CNA43, DNA43, PRO2249 | ||
| Description | minichromosome maintenance complex component 10 | ||
| EnsEMBL link | ENSG00000065328 | ||
| Chromosome | 10 - forward strand | ||
| Chromosomal location (UCSC link) |
13203554 - 13253095 (49542bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_018518 NM_182751 AK292701 AK055695 BC101727 AB042719 AL136840 BC143490 BC009108 BC004876 AF119869 CR620605 |
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 5 | 3 | NM_018518 AB042719 AL136840 BC143490 |
|
| Intron retentions |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| IR1 | 19 | AL136840 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 13203554 | NM_018518 NM_182751 |
| P2 | 1 | 26 | 13203579 | AK292701 |
| P3 | 1 | 28 | 13203581 | AK055695 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_018518 | sequence |
| NM_182751 | sequence |
| AK292701 | sequence |
| AK055695 | sequence |
| BC101727 | sequence |
| AB042719 | sequence |
| AL136840 | sequence |
| BC143490 | sequence |
| BC009108 | sequence |
| BC004876 | sequence |
| AF119869 | sequence |
| CR620605 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 15 | 36132 | 13239685 | no | AK055695 | Full_length | |
| PA2 | 20 | 48183 | 13251736 | no | AK292701 | Full_length | |
| PA3 | 20 | 48345 | 13251898 | ATTAAA | no | AA583783 | 3prime_est |
| PA4 | 20 | 49542 | 13253095 | yes | NM_018518 | Cdna | |
| PA5 | 20 | 49542 | 13253095 | yes | NM_182751 | Cdna | |
| PA6 | 20 | 49542 | 13253095 | yes | AL136840 | Cdna | |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 3 |
| Transcription termination & last exon(s) | |
| 15 | 1 |
| 20 | 5 |
| Alternative 3' splice sites | |
| 5 | 1 |
| Intron retentions | |
| 19 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| 16 | Expression | Expression | Expression |
| 17 | Expression | Expression | Expression |
| 18 | Expression | Expression | Expression |
| 19 | Expression | Expression | Expression |
| 20 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 53 | 13203554 - 13203606 | 53 | sequence | download |
| 1 | Intron | 54 - 2574 | 13203607 - 13206127 | 2521 | sequence | - |
| 2 | Exon | 2575 - 2656 | 13206128 - 13206209 | 82 | sequence | download |
| 2 | Intron | 2657 - 9368 | 13206210 - 13212921 | 6712 | sequence | - |
| 3 | Exon | 9369 - 9710 | 13212922 - 13213263 | 342 | sequence | download |
| 3 | Intron | 9711 - 10822 | 13213264 - 13214375 | 1112 | sequence | - |
| 4 | Exon | 10823 - 10927 | 13214376 - 13214480 | 105 | sequence | download |
| 4 | Intron | 10928 - 11071 | 13214481 - 13214624 | 144 | sequence | - |
| 5 | Exon | 11072 - 11212 | 13214625 - 13214765 | 141 | sequence | download |
| 5 | Intron | 11213 - 13956 | 13214766 - 13217509 | 2744 | sequence | - |
| 6 | Exon | 13957 - 14128 | 13217510 - 13217681 | 172 | sequence | download |
| 6 | Intron | 14129 - 18888 | 13217682 - 13222441 | 4760 | sequence | - |
| 7 | Exon | 18889 - 19054 | 13222442 - 13222607 | 166 | sequence | download |
| 7 | Intron | 19055 - 21379 | 13222608 - 13224932 | 2325 | sequence | - |
| 8 | Exon | 21380 - 21547 | 13224933 - 13225100 | 168 | sequence | download |
| 8 | Intron | 21548 - 24610 | 13225101 - 13228163 | 3063 | sequence | - |
| 9 | Exon | 24611 - 24727 | 13228164 - 13228280 | 117 | sequence | download |
| 9 | Intron | 24728 - 27327 | 13228281 - 13230880 | 2600 | sequence | - |
| 10 | Exon | 27328 - 27527 | 13230881 - 13231080 | 200 | sequence | download |
| 10 | Intron | 27528 - 29745 | 13231081 - 13233298 | 2218 | sequence | - |
| 11 | Exon | 29746 - 29846 | 13233299 - 13233399 | 101 | sequence | download |
| 11 | Intron | 29847 - 30701 | 13233400 - 13234254 | 855 | sequence | - |
| 12 | Exon | 30702 - 30812 | 13234255 - 13234365 | 111 | sequence | download |
| 12 | Intron | 30813 - 30897 | 13234366 - 13234450 | 85 | sequence | - |
| 13 | Exon | 30898 - 31015 | 13234451 - 13234568 | 118 | sequence | download |
| 13 | Intron | 31016 - 33487 | 13234569 - 13237040 | 2472 | sequence | - |
| 14 | Exon | 33488 - 33716 | 13237041 - 13237269 | 229 | sequence | download |
| 14 | Intron | 33717 - 36069 | 13237270 - 13239622 | 2353 | sequence | - |
| 15 | Exon | 36070 - 36214 | 13239623 - 13239767 | 145 | sequence | download |
| 15 | Intron | 36215 - 37135 | 13239768 - 13240688 | 921 | sequence | - |
| 16 | Exon | 37136 - 37254 | 13240689 - 13240807 | 119 | sequence | download |
| 16 | Intron | 37255 - 39867 | 13240808 - 13243420 | 2613 | sequence | - |
| 17 | Exon | 39868 - 39981 | 13243421 - 13243534 | 114 | sequence | download |
| 17 | Intron | 39982 - 42665 | 13243535 - 13246218 | 2684 | sequence | - |
| 18 | Exon | 42666 - 42811 | 13246219 - 13246364 | 146 | sequence | download |
| 18 | Intron | 42812 - 47537 | 13246365 - 13251090 | 4726 | sequence | - |
| 19 | Exon | 47538 - 47580 | 13251091 - 13251133 | 43 | sequence | download |
| 19 | Intron | 47581 - 47673 | 13251134 - 13251226 | 93 | sequence | - |
| 20 | Exon | 47674 - 49542 | 13251227 - 13253095 | 1869 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-384 | 47658 - 47677 | 13251211 - 13251230 | miranda |
| Group 2 | ||||
| M1 | hsa-miR-194 | 47693 - 47714 | 13251246 - 13251267 | miranda |
| Group 3 | ||||
| M1 | hsa-miR-339-5p | 48506 - 48528 | 13252059 - 13252081 | miranda |
| M2 | hsa-miR-590-3p | 49037 - 49056 | 13252590 - 13252609 | miranda |
| M3 | hsa-miR-340 | 49155 - 49178 | 13252708 - 13252731 | miranda |
| M4 | hsa-miR-653 | 49190 - 49211 | 13252743 - 13252764 | miranda |
| M5 | hsa-miR-373 | 49231 - 49255 | 13252784 - 13252808 | miranda |
| M6 | hsa-miR-372 | 49233 - 49255 | 13252786 - 13252808 | miranda |
| M7 | hsa-miR-520e | 49233 - 49255 | 13252786 - 13252808 | miranda |
| M8 | hsa-miR-302a | 49233 - 49255 | 13252786 - 13252808 | miranda |
| M9 | hsa-miR-302b | 49233 - 49255 | 13252786 - 13252808 | miranda |
| M10 | hsa-miR-302d | 49233 - 49255 | 13252786 - 13252808 | miranda |
| M11 | hsa-miR-302c | 49234 - 49255 | 13252787 - 13252808 | miranda |
| M12 | hsa-miR-520c-3p | 49234 - 49255 | 13252787 - 13252808 | miranda |
| M13 | hsa-miR-520b | 49235 - 49255 | 13252788 - 13252808 | miranda |
| M14 | hsa-miR-520a-3p | 49237 - 49255 | 13252790 - 13252808 | miranda |
| M15 | hsa-miR-520d-3p | 49237 - 49255 | 13252790 - 13252808 | miranda |
| M16 | hsa-miR-302e | 49240 - 49255 | 13252793 - 13252808 | miranda |
| M17 | hsa-miR-410 | 49437 - 49456 | 13252990 - 13253009 | miranda |










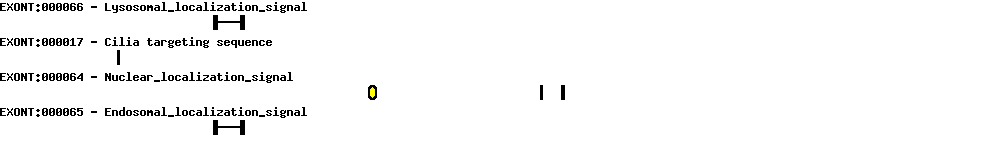



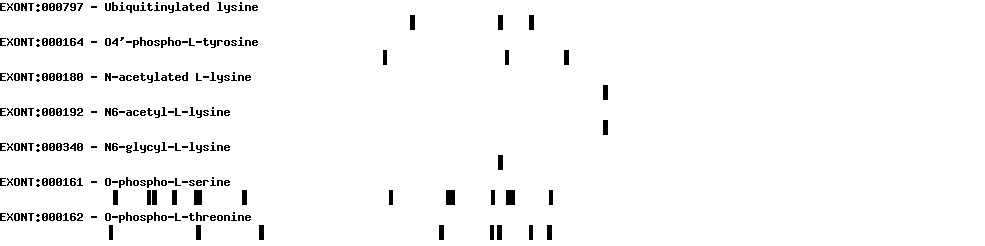


Please wait while loading ...