
| Gene |  |
||
|---|---|---|---|
| Symbol | CAMK1D | ||
| Synonyms | CKLiK | ||
| Description | calcium/calmodulin-dependent protein kinase ID | ||
| EnsEMBL link | ENSG00000183049 | ||
| Chromosome | 10 - forward strand | ||
| Chromosomal location (UCSC link) |
12391583 - 12871735 (480153bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_020397 NM_153498 AB081726 CR616818 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 12391583 | NM_020397 NM_153498 |
| P2 | 3 | 317153 | 12708735 | CR616818 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_020397 | sequence |
| NM_153498 | sequence |
| AB081726 | sequence |
| CR616818 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 10 | 476560 | 12868142 | no | NM_020397 | Cdna | |
| PA2 | 11 | 480153 | 12871735 | no | NM_153498 | Cdna | |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 9784 | 2 | 2 |
| 9784 | 4 | 3 |
| 9784 | 5 | 4 |
| 9784 | 6 | 5 |
| 9784 | 7 | 6 |
| 9784 | 8 | 7 |
| 9784 | 9 | 8 |
| 9784 | 10 | 9 |
| 9784 | 11 | 10 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 1 |
| 3 | 1 |
| Transcription termination & last exon(s) | |
| 10 | 1 |
| 11 | 1 |
| Exon skipping | |
| 3 | 3 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 327 | 12391583 - 12391909 | 327 | sequence | download |
| 1 | Intron | 328 - 203641 | 12391910 - 12595223 | 203314 | sequence | - |
| 2 | Exon | 203642 - 203774 | 12595224 - 12595356 | 133 | sequence | download |
| 2 | Intron | 203775 - 317152 | 12595357 - 12708734 | 113378 | sequence | - |
| 3 | Exon | 317153 - 317227 | 12708735 - 12708809 | 75 | sequence | download |
| 3 | Intron | 317228 - 411364 | 12708810 - 12802946 | 94137 | sequence | - |
| 4 | Exon | 411365 - 411503 | 12802947 - 12803085 | 139 | sequence | download |
| 4 | Intron | 411504 - 420089 | 12803086 - 12811671 | 8586 | sequence | - |
| 5 | Exon | 420090 - 420216 | 12811672 - 12811798 | 127 | sequence | download |
| 5 | Intron | 420217 - 441574 | 12811799 - 12833156 | 21358 | sequence | - |
| 6 | Exon | 441575 - 441650 | 12833157 - 12833232 | 76 | sequence | download |
| 6 | Intron | 441651 - 464611 | 12833233 - 12856193 | 22961 | sequence | - |
| 7 | Exon | 464612 - 464724 | 12856194 - 12856306 | 113 | sequence | download |
| 7 | Intron | 464725 - 466666 | 12856307 - 12858248 | 1942 | sequence | - |
| 8 | Exon | 466667 - 466745 | 12858249 - 12858327 | 79 | sequence | download |
| 8 | Intron | 466746 - 474881 | 12858328 - 12866463 | 8136 | sequence | - |
| 9 | Exon | 474882 - 474969 | 12866464 - 12866551 | 88 | sequence | download |
| 9 | Intron | 474970 - 475989 | 12866552 - 12867571 | 1020 | sequence | - |
| 10 | Exon | 475990 - 476107 | 12867572 - 12867689 | 118 | sequence | download |
| 10 | Intron | 476108 - 479185 | 12867690 - 12870767 | 3078 | sequence | - |
| 11 | Exon | 479186 - 480153 | 12870768 - 12871735 | 968 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-767-5p | 479398 - 479405 | 12870980 - 12870987 | pita |
| M2 | hsa-miR-145 | 479421 - 479428 | 12871003 - 12871010 | targetscan |
| M3 | hsa-miR-181d | 479454 - 479476 | 12871036 - 12871058 | miranda |
| M4 | hsa-miR-936 | 479455 - 479461 | 12871037 - 12871043 | pita |
| M5 | hsa-miR-367 | 479460 - 479467 | 12871042 - 12871049 | pictar |
| M6 | hsa-miR-539 | 479463 - 479485 | 12871045 - 12871067 | miranda |
| M7 | hsa-miR-223 | 479474 - 479496 | 12871056 - 12871078 | miranda |
| M8 | hsa-miR-539 | 479478 - 479484 | 12871060 - 12871066 | pita |
| M9 | hsa-miR-539 | 479478 - 479486 | 12871060 - 12871068 | targetscan |
| M10 | hsa-miR-758 | 479483 - 479490 | 12871065 - 12871072 | targetscan |
| M11 | hsa-miR-582-5p | 479488 - 479494 | 12871070 - 12871076 | pita |
| M12 | hsa-miR-29b | 479526 - 479532 | 12871108 - 12871114 | pita |
| M13 | hsa-miR-29a | 479526 - 479533 | 12871108 - 12871115 | targetscan |
| M14 | hsa-miR-29c | 479526 - 479532 | 12871108 - 12871114 | pita |
| M15 | hsa-miR-29b | 479526 - 479533 | 12871108 - 12871115 | targetscan |
| M16 | hsa-miR-29c | 479526 - 479533 | 12871108 - 12871115 | pictar |
| M17 | hsa-miR-29c | 479526 - 479533 | 12871108 - 12871115 | targetscan |
| M18 | hsa-miR-29b | 479526 - 479533 | 12871108 - 12871115 | pictar |
| M19 | hsa-miR-29a | 479526 - 479533 | 12871108 - 12871115 | pictar |
| M20 | hsa-miR-29a | 479526 - 479532 | 12871108 - 12871114 | pita |
| M21 | hsa-miR-31 | 479543 - 479550 | 12871125 - 12871132 | pictar |
| M22 | hsa-miR-450b-5p | 479546 - 479552 | 12871128 - 12871134 | pita |
| M23 | hsa-miR-1253 | 479606 - 479612 | 12871188 - 12871194 | pita |
| M24 | hsa-miR-1300 | 479607 - 479613 | 12871189 - 12871195 | pita |
| M25 | hsa-miR-329 | 479614 - 479639 | 12871196 - 12871221 | miranda |
| M26 | hsa-miR-362-3p | 479619 - 479639 | 12871201 - 12871221 | miranda |
| M27 | hsa-miR-409-5p | 479619 - 479625 | 12871201 - 12871207 | pita |
| M28 | hsa-miR-329 | 479632 - 479638 | 12871214 - 12871220 | pita |
| M29 | hsa-miR-362-3p | 479632 - 479638 | 12871214 - 12871220 | pita |
| M30 | hsa-miR-145 | 479656 - 479678 | 12871238 - 12871260 | miranda |
| M31 | hsa-miR-145 | 479671 - 479678 | 12871253 - 12871260 | pictar |
| M32 | hsa-miR-145 | 479671 - 479678 | 12871253 - 12871260 | targetscan |
| M33 | hsa-miR-145 | 479671 - 479677 | 12871253 - 12871259 | pita |
| M34 | hsa-miR-495 | 479696 - 479718 | 12871278 - 12871300 | miranda |
| M35 | hsa-miR-338-5p | 479696 - 479702 | 12871278 - 12871284 | pita |
| M36 | hsa-miR-367 | 479701 - 479708 | 12871283 - 12871290 | pictar |
| M37 | hsa-miR-606 | 479784 - 479790 | 12871366 - 12871372 | pita |
| M38 | hsa-miR-216b | 479791 - 479813 | 12871373 - 12871395 | miranda |
| M39 | hsa-miR-590-3p | 479835 - 479855 | 12871417 - 12871437 | miranda |
| M40 | hsa-miR-590-3p | 479848 - 479854 | 12871430 - 12871436 | pita |
| M41 | hsa-miR-29c | 479874 - 479881 | 12871456 - 12871463 | pictar |
| M42 | hsa-miR-29b | 479874 - 479881 | 12871456 - 12871463 | pictar |
| M43 | hsa-miR-29a | 479874 - 479881 | 12871456 - 12871463 | pictar |
| M44 | hsa-miR-367 | 479875 - 479882 | 12871457 - 12871464 | pictar |


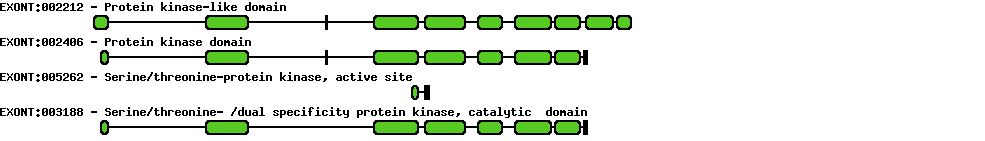





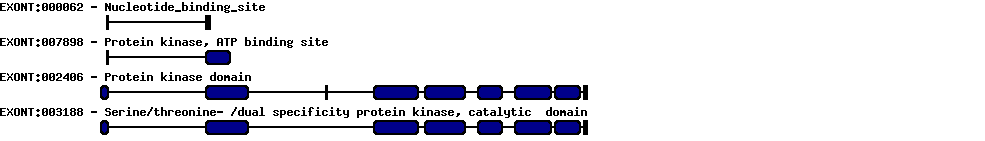



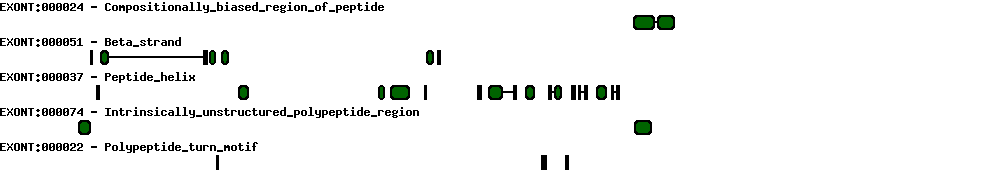

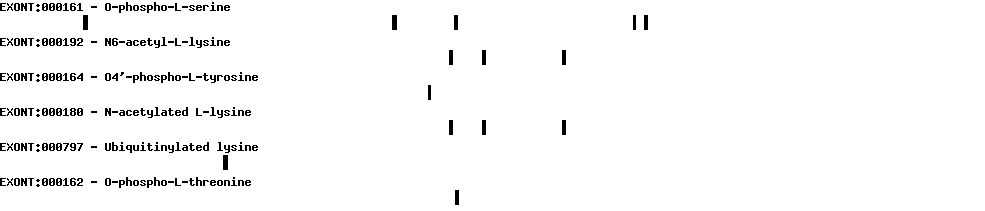


Please wait while loading ...