
 FasterDB identifier: 16790 - Dmc1 (Dmc1, Dmc1h, Mei11, sgdp)
DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast) Gene
FasterDB identifier: 16790 - Dmc1 (Dmc1, Dmc1h, Mei11, sgdp)
DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast) Gene

| Gene |  |
||
|---|---|---|---|
| Symbol | Dmc1 | ||
| Synonyms | Dmc1, Dmc1h, Mei11, sgdp | ||
| Description | DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast) Gene | ||
| EnsEMBL link | ENSMUSG00000022429 | ||
| Chromosome | 15 - reverse strand | ||
| Chromosomal location (UCSC link) |
79391930 - 79435539 (43610bp) | ||
| Sequence | sequence | ||
| Associated transcripts | AK169570 AK076777 D58419 D64107 NM_010059 BC116767 BC119081 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 79435539 | AK169570 |
| P2 | 1 | 10 | 79435530 | AK076777 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| AK169570 | sequence |
| AK076777 | sequence |
| D58419 | sequence |
| D64107 | sequence |
| NM_010059 | sequence |
| BC116767 | sequence |
| BC119081 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 14 | 43610 | 79391930 | AGTAAA | no | D64107 | Cdna |
| PA2 | 14 | 43610 | 79391930 | AGTAAA | no | NM_010059 | Cdna |
| Exon skipping |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ES1 | 6 | AK169570 |
| ES2 | 7 | AK169570 |
| ES3 | 8 | AK169570 AK076777 |
| ES4 | 9 | AK169570 AK076777 |
| ES5 | 11 | AK169570 |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier human | Position mouse | Position human |
| 2289 | 1 | 1 |
| 2289 | 2 | 2 |
| 2289 | 3 | 3 |
| 2289 | 4 | 4 |
| 2289 | 5 | 5 |
| 2289 | 6 | 6 |
| 2289 | 7 | 7 |
| 2289 | 8 | 8 |
| 2289 | 9 | 10 |
| 2289 | 10 | 11 |
| 2289 | 11 | 12 |
| 2289 | 12 | 13 |
| 2289 | 13 | 14 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 2 |
| Transcription termination & last exon(s) | |
| 14 | 2 |
| Exon skipping | |
| 6 | 1 |
| 7 | 1 |
| 8 | 2 |
| 9 | 2 |
| 11 | 1 |
| Expression |  |
|
|---|---|---|
| Exon position | Tissues | Cell lines |
| 1 | Expression | Expression |
| 2 | Expression | Expression |
| 3 | Expression | Expression |
| 4 | Expression | Expression |
| 5 | Expression | Expression |
| 6 | Expression | Expression |
| 7 | Expression | Expression |
| 8 | Expression | Expression |
| 9 | Expression | Expression |
| 10 | Expression | Expression |
| 11 | Expression | Expression |
| 12 | Expression | Expression |
| 13 | Expression | Expression |
| 14 | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 148 | 79435392-79435539 | 148 | sequence | download |
| 1 | Intron | 149 - 2625 | 79432915-79435391 | 2477 | sequence | - |
| 2 | Exon | 2626 - 2709 | 79432831-79432914 | 84 | sequence | download |
| 2 | Intron | 2710 - 3548 | 79431992-79432830 | 839 | sequence | - |
| 3 | Exon | 3549 - 3593 | 79431947-79431991 | 45 | sequence | download |
| 3 | Intron | 3594 - 4917 | 79430623-79431946 | 1324 | sequence | - |
| 4 | Exon | 4918 - 5064 | 79430476-79430622 | 147 | sequence | download |
| 4 | Intron | 5065 - 8783 | 79426757-79430475 | 3719 | sequence | - |
| 5 | Exon | 8784 - 8866 | 79426674-79426756 | 83 | sequence | download |
| 5 | Intron | 8867 - 11922 | 79423618-79426673 | 3056 | sequence | - |
| 6 | Exon | 11923 - 11975 | 79423565-79423617 | 53 | sequence | download |
| 6 | Intron | 11976 - 13463 | 79422077-79423564 | 1488 | sequence | - |
| 7 | Exon | 13464 - 13505 | 79422035-79422076 | 42 | sequence | download |
| 7 | Intron | 13506 - 16286 | 79419254-79422034 | 2781 | sequence | - |
| 8 | Exon | 16287 - 16359 | 79419181-79419253 | 73 | sequence | download |
| 8 | Intron | 16360 - 19349 | 79416191-79419180 | 2990 | sequence | - |
| 9 | Exon | 19350 - 19441 | 79416099-79416190 | 92 | sequence | download |
| 9 | Intron | 19442 - 19938 | 79415602-79416098 | 497 | sequence | - |
| 10 | Exon | 19939 - 20012 | 79415528-79415601 | 74 | sequence | download |
| 10 | Intron | 20013 - 20133 | 79415407-79415527 | 121 | sequence | - |
| 11 | Exon | 20134 - 20248 | 79415292-79415406 | 115 | sequence | download |
| 11 | Intron | 20249 - 20730 | 79414810-79415291 | 482 | sequence | - |
| 12 | Exon | 20731 - 20791 | 79414749-79414809 | 61 | sequence | download |
| 12 | Intron | 20792 - 36354 | 79399186-79414748 | 15563 | sequence | - |
| 13 | Exon | 36355 - 36471 | 79399069-79399185 | 117 | sequence | download |
| 13 | Intron | 36472 - 42527 | 79393013-79399068 | 6056 | sequence | - |
| 14 | Exon | 42528 - 43610 | 79391930-79393012 | 1083 | sequence | download |
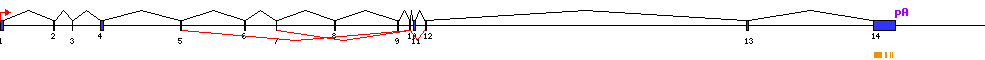
Please wait while loading ...