
 FasterDB identifier: 2202 - TMEM184B (C22orf5, DKFZP586A1024, FM08, HS5O6A)
transmembrane protein 184B
FasterDB identifier: 2202 - TMEM184B (C22orf5, DKFZP586A1024, FM08, HS5O6A)
transmembrane protein 184B

| Gene |  |
||
|---|---|---|---|
| Symbol | TMEM184B | ||
| Synonyms | C22orf5, DKFZP586A1024, FM08, HS5O6A | ||
| Description | transmembrane protein 184B | ||
| EnsEMBL link | ENSG00000198792 | ||
| Chromosome | 22 - reverse strand | ||
| Chromosomal location (UCSC link) |
38615298 - 38669040 (53743bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_001195072 NM_012264 AK292652 AB097053 AK309071 BC015489 CR749859 NM_001195071 BX648345 CR456413 AL096879 AK075417 AK309607 BC012983 |
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 10 | 65 | NM_001195072 NM_012264 AK292652 AB097053 BC015489 CR749859 NM_001195071 BX648345 CR456413 AL096879 AK075417 |
|
| Alternative 5' splice sites (donor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 12 | -25 | AK309071 |
|
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 38669040 | NM_001195072 NM_012264 AK292652 |
| P2 | 1 | 30 | 38669011 | AK309071 |
| P3 | 2 | 371 | 38668670 | NM_001195071 BX648345 |
| P4 | 6 | 29247 | 38639794 | AK309607 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_001195072 | sequence |
| NM_012264 | sequence |
| AK292652 | sequence |
| AB097053 | sequence |
| AK309071 | sequence |
| BC015489 | sequence |
| CR749859 | sequence |
| NM_001195071 | sequence |
| BX648345 | sequence |
| CR456413 | sequence |
| AL096879 | sequence |
| AK075417 | sequence |
| AK309607 | sequence |
| BC012983 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 9 | 33665 | 38635376 | no | AK309607 | Full_length | |
| PA2 | 13 | 47607 | 38621434 | no | AK309071 | Full_length | |
| PA3 | 15 | 51684 | 38617357 | no | AK292652 | Full_length | |
| PA4 | 15 | 53559 | 38615482 | CATAAA | yes | BM127731 | 3prime_est |
| PA5 | 15 | 53743 | 38615298 | AATATA | yes | NM_001195072 | Cdna |
| PA6 | 15 | 53743 | 38615298 | AATATA | yes | NM_012264 | Cdna |
| PA7 | 15 | 53743 | 38615298 | AATATA | yes | NM_001195071 | Cdna |
| PA8 | 15 | 53743 | 38615298 | AATATA | no | AL096879 | Cdna |
| PA9 | 15 | 53743 | 38615298 | AATATA | no | AK075417 | Cdna |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 16406 | 3 | 3 |
| 16406 | 5 | 4 |
| 16406 | 10 | 5 |
| 16406 | 11 | 6 |
| 16406 | 12 | 7 |
| 16406 | 13 | 8 |
| 16406 | 14 | 9 |
| 16406 | 15 | 11 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 2 |
| 2 | 1 |
| 6 | 1 |
| Transcription termination & last exon(s) | |
| 9 | 1 |
| 13 | 1 |
| 15 | 7 |
| Exon skipping | |
| 2 | 7 |
| 4 | 9 |
| 6 | 12 |
| 7 | 12 |
| 8 | 12 |
| 9 | 12 |
| Alternative 3' splice sites | |
| 10 | 1 |
| Alternative 5' splice sites | |
| 12 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 151 | 38668890-38669040 | 151 | sequence | download |
| 1 | Intron | 152 - 370 | 38668671-38668889 | 219 | sequence | - |
| 2 | Exon | 371 - 536 | 38668505-38668670 | 166 | sequence | download |
| 2 | Intron | 537 - 25015 | 38644026-38668504 | 24479 | sequence | - |
| 3 | Exon | 25016 - 25265 | 38643776-38644025 | 250 | sequence | download |
| 3 | Intron | 25266 - 26149 | 38642892-38643775 | 884 | sequence | - |
| 4 | Exon | 26150 - 26250 | 38642791-38642891 | 101 | sequence | download |
| 4 | Intron | 26251 - 26934 | 38642107-38642790 | 684 | sequence | - |
| 5 | Exon | 26935 - 27100 | 38641941-38642106 | 166 | sequence | download |
| 5 | Intron | 27101 - 29246 | 38639795-38641940 | 2146 | sequence | - |
| 6 | Exon | 29247 - 29776 | 38639265-38639794 | 530 | sequence | download |
| 6 | Intron | 29777 - 30802 | 38638239-38639264 | 1026 | sequence | - |
| 7 | Exon | 30803 - 30827 | 38638214-38638238 | 25 | sequence | download |
| 7 | Intron | 30828 - 31410 | 38637631-38638213 | 583 | sequence | - |
| 8 | Exon | 31411 - 31683 | 38637358-38637630 | 273 | sequence | download |
| 8 | Intron | 31684 - 33308 | 38635733-38637357 | 1625 | sequence | - |
| 9 | Exon | 33309 - 33665 | 38635376-38635732 | 357 | sequence | download |
| 9 | Intron | 33666 - 41635 | 38627406-38635375 | 7970 | sequence | - |
| 10 | Exon | 41636 - 41791 | 38627250-38627405 | 156 | sequence | download |
| 10 | Intron | 41792 - 42290 | 38626751-38627249 | 499 | sequence | - |
| 11 | Exon | 42291 - 42366 | 38626675-38626750 | 76 | sequence | download |
| 11 | Intron | 42367 - 46163 | 38622878-38626674 | 3797 | sequence | - |
| 12 | Exon | 46164 - 46255 | 38622786-38622877 | 92 | sequence | download |
| 12 | Intron | 46256 - 47440 | 38621601-38622785 | 1185 | sequence | - |
| 13 | Exon | 47441 - 47610 | 38621431-38621600 | 170 | sequence | download |
| 13 | Intron | 47611 - 48054 | 38620987-38621430 | 444 | sequence | - |
| 14 | Exon | 48055 - 48249 | 38620792-38620986 | 195 | sequence | download |
| 14 | Intron | 48250 - 51323 | 38617718-38620791 | 3074 | sequence | - |
| 15 | Exon | 51324 - 53743 | 38615298-38617717 | 2420 | sequence | download |

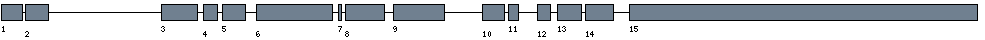

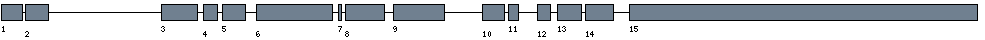

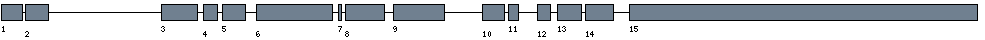

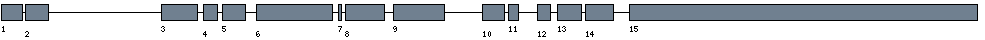

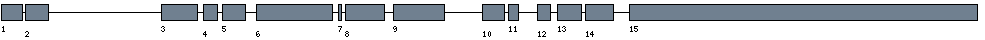




Please wait while loading ...