
| Gene |  |
||
|---|---|---|---|
| Symbol | SMURF2 | ||
| Synonyms | ENSG00000108854 | ||
| Description | SMAD specific E3 ubiquitin protein ligase 2 | ||
| EnsEMBL link | ENSG00000108854 | ||
| Chromosome | 17 - reverse strand | ||
| Chromosomal location (UCSC link) |
62538418 - 62658386 (119969bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_022739 CR936721 AY014180 BC051283 BC093876 BC111945 AF301463 AF310676 BC009527 |
| Exon deletions |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ED1 | 1 | CR936721 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 62658386 | NM_022739 |
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_022739 | sequence |
| CR936721 | sequence |
| AY014180 | sequence |
| BC051283 | sequence |
| BC093876 | sequence |
| BC111945 | sequence |
| AF301463 | sequence |
| AF310676 | sequence |
| BC009527 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 20 | 119969 | 62538418 | yes | CR936721 | Cdna | |
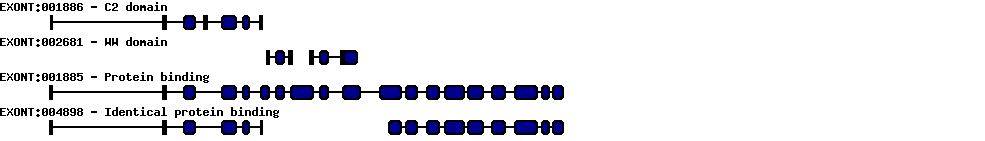
| Exon skipping |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ES1 | 2 | CR936721 AF301463 AF310676 |
| ES2 | 4 | NM_022739 AY014180 BC093876 BC111945 AF301463 AF310676 |
| ES3 | 7 | BC051283 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 1 |
| Transcription termination & last exon(s) | |
| 20 | 1 |
| Exon skipping | |
| 2 | 3 |
| 4 | 6 |
| 7 | 1 |
| Exon deletions | |
| 1 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| 16 | Expression | Expression | Expression |
| 17 | Expression | Expression | Expression |
| 18 | Expression | Expression | Expression |
| 19 | Expression | Expression | Expression |
| 20 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 440 | 62657947-62658386 | 440 | sequence | download |
| 1 | Intron | 441 - 55628 | 62602759-62657946 | 55188 | sequence | - |
| 2 | Exon | 55629 - 55667 | 62602720-62602758 | 39 | sequence | download |
| 2 | Intron | 55668 - 63778 | 62594609-62602719 | 8111 | sequence | - |
| 3 | Exon | 63779 - 63887 | 62594500-62594608 | 109 | sequence | download |
| 3 | Intron | 63888 - 68164 | 62590223-62594499 | 4277 | sequence | - |
| 4 | Exon | 68165 - 68278 | 62590109-62590222 | 114 | sequence | download |
| 4 | Intron | 68279 - 68695 | 62589692-62590108 | 417 | sequence | - |
| 5 | Exon | 68696 - 68829 | 62589558-62589691 | 134 | sequence | download |
| 5 | Intron | 68830 - 71119 | 62587268-62589557 | 2290 | sequence | - |
| 6 | Exon | 71120 - 71185 | 62587202-62587267 | 66 | sequence | download |
| 6 | Intron | 71186 - 76098 | 62582289-62587201 | 4913 | sequence | - |
| 7 | Exon | 76099 - 76183 | 62582204-62582288 | 85 | sequence | download |
| 7 | Intron | 76184 - 78724 | 62579663-62582203 | 2541 | sequence | - |
| 8 | Exon | 78725 - 78808 | 62579579-62579662 | 84 | sequence | download |
| 8 | Intron | 78809 - 81277 | 62577110-62579578 | 2469 | sequence | - |
| 9 | Exon | 81278 - 81480 | 62576907-62577109 | 203 | sequence | download |
| 9 | Intron | 81481 - 83692 | 62574695-62576906 | 2212 | sequence | - |
| 10 | Exon | 83693 - 83777 | 62574610-62574694 | 85 | sequence | download |
| 10 | Intron | 83778 - 90312 | 62568075-62574609 | 6535 | sequence | - |
| 11 | Exon | 90313 - 90471 | 62567916-62568074 | 159 | sequence | download |
| 11 | Intron | 90472 - 99302 | 62559085-62567915 | 8831 | sequence | - |
| 12 | Exon | 99303 - 99498 | 62558889-62559084 | 196 | sequence | download |
| 12 | Intron | 99499 - 100665 | 62557722-62558888 | 1167 | sequence | - |
| 13 | Exon | 100666 - 100769 | 62557618-62557721 | 104 | sequence | download |
| 13 | Intron | 100770 - 104546 | 62553841-62557617 | 3777 | sequence | - |
| 14 | Exon | 104547 - 104661 | 62553726-62553840 | 115 | sequence | download |
| 14 | Intron | 104662 - 106270 | 62552117-62553725 | 1609 | sequence | - |
| 15 | Exon | 106271 - 106449 | 62551938-62552116 | 179 | sequence | download |
| 15 | Intron | 106450 - 107275 | 62551112-62551937 | 826 | sequence | - |
| 16 | Exon | 107276 - 107413 | 62550974-62551111 | 138 | sequence | download |
| 16 | Intron | 107414 - 110564 | 62547823-62550973 | 3151 | sequence | - |
| 17 | Exon | 110565 - 110685 | 62547702-62547822 | 121 | sequence | download |
| 17 | Intron | 110686 - 114467 | 62543920-62547701 | 3782 | sequence | - |
| 18 | Exon | 114468 - 114669 | 62543718-62543919 | 202 | sequence | download |
| 18 | Intron | 114670 - 115930 | 62542457-62543717 | 1261 | sequence | - |
| 19 | Exon | 115931 - 116006 | 62542381-62542456 | 76 | sequence | download |
| 19 | Intron | 116007 - 116321 | 62542066-62542380 | 315 | sequence | - |
| 20 | Exon | 116322 - 119969 | 62538418-62542065 | 3648 | sequence | download |

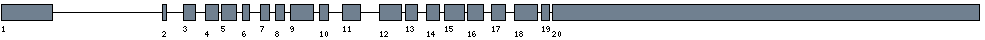



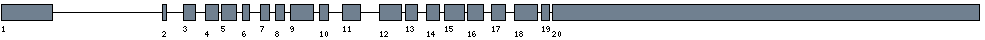
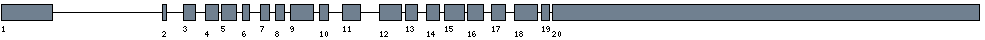
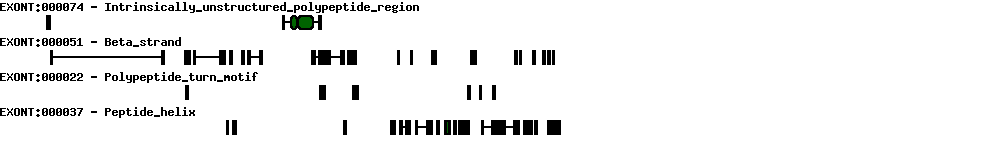
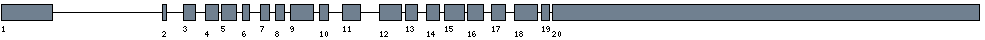
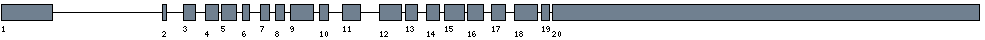

Please wait while loading ...