
| Gene |  |
||
|---|---|---|---|
| Symbol | PHYH | ||
| Synonyms | PAHX, PHYH1, RD | ||
| Description | phytanoyl-CoA 2-hydroxylase | ||
| EnsEMBL link | ENSG00000107537 | ||
| Chromosome | 10 - reverse strand | ||
| Chromosomal location (UCSC link) |
13319796 - 13344412 (24617bp) | ||
| Sequence | sequence | ||
| Associated transcripts | CR623416 NM_006214 AF112977 BC029512 CR610525 CR600223 AF023462 CR597553 CR591194 CR623967 CR542055 NM_001037537 |
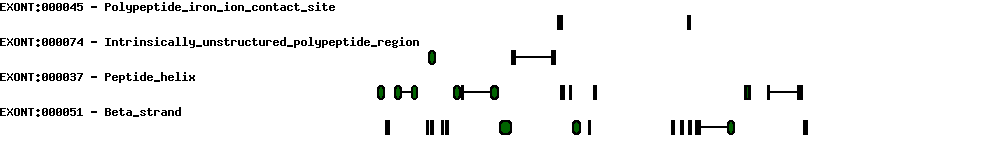
| Alternative 3' splice site (acceptor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 4 | 4 | CR623416 |
|
| Alternative 5' splice sites (donor) |  |
|||
|---|---|---|---|---|
| Identifier | Position | Distance (bp) | Transcript | |
| ASS1 | 7 | -6 | NM_006214 AF112977 BC029512 CR610525 CR600223 AF023462 CR597553 CR591194 CR623967 CR542055 NM_001037537 |
|
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 13344412 | CR623416 |
| P2 | 2 | 2283 | 13342130 | NM_006214 |
| P3 | 3 | 2667 | 13341746 | NM_001037537 |
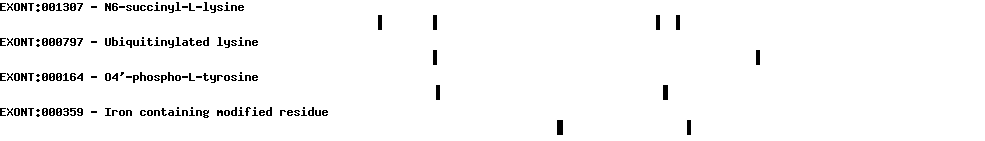
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| CR623416 | sequence |
| NM_006214 | sequence |
| AF112977 | sequence |
| BC029512 | sequence |
| CR610525 | sequence |
| CR600223 | sequence |
| AF023462 | sequence |
| CR597553 | sequence |
| CR591194 | sequence |
| CR623967 | sequence |
| CR542055 | sequence |
| NM_001037537 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 11 | 24375 | 13320038 | AATAAA | no | AW473079 | 3prime_est |
| PA2 | 11 | 24617 | 13319796 | AATAAA | no | CR610525 | Cdna |
| PA3 | 11 | 24617 | 13319796 | AATAAA | no | CR623967 | Cdna |
| PA4 | 11 | 25510 | 13318903 | AATAAA | no | BE645857 | 3prime_est |
| Exon skipping |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ES1 | 2 | CR623416 |
| ES2 | 3 | CR623416 NM_006214 AF112977 BC029512 CR610525 CR600223 AF023462 CR597553 CR591194 CR623967 CR542055 |
| ES3 | 4 | NM_001037537 |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 9279 | 4 | 2 |
| 9279 | 5 | 3 |
| 9279 | 6 | 4 |
| 9279 | 7 | 5 |
| 9279 | 8 | 6 |
| 9279 | 9 | 7 |
| 9279 | 10 | 8 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 1 |
| 2 | 1 |
| 3 | 1 |
| Transcription termination & last exon(s) | |
| 11 | 4 |
| Exon skipping | |
| 2 | 1 |
| 3 | 11 |
| 4 | 1 |
| Alternative 3' splice sites | |
| 4 | 1 |
| Alternative 5' splice sites | |
| 7 | 1 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 427 | 13343986-13344412 | 427 | sequence | download |
| 1 | Intron | 428 - 2282 | 13342131-13343985 | 1855 | sequence | - |
| 2 | Exon | 2283 - 2445 | 13341968-13342130 | 163 | sequence | download |
| 2 | Intron | 2446 - 2666 | 13341747-13341967 | 221 | sequence | - |
| 3 | Exon | 2667 - 2993 | 13341420-13341746 | 327 | sequence | download |
| 3 | Intron | 2994 - 4167 | 13340246-13341419 | 1174 | sequence | - |
| 4 | Exon | 4168 - 4226 | 13340187-13340245 | 59 | sequence | download |
| 4 | Intron | 4227 - 6806 | 13337607-13340186 | 2580 | sequence | - |
| 5 | Exon | 6807 - 6917 | 13337496-13337606 | 111 | sequence | download |
| 5 | Intron | 6918 - 7816 | 13336597-13337495 | 899 | sequence | - |
| 6 | Exon | 7817 - 7985 | 13336428-13336596 | 169 | sequence | download |
| 6 | Intron | 7986 - 10500 | 13333913-13336427 | 2515 | sequence | - |
| 7 | Exon | 10501 - 10588 | 13333825-13333912 | 88 | sequence | download |
| 7 | Intron | 10589 - 13871 | 13330542-13333824 | 3283 | sequence | - |
| 8 | Exon | 13872 - 14053 | 13330360-13330541 | 182 | sequence | download |
| 8 | Intron | 14054 - 18573 | 13325840-13330359 | 4520 | sequence | - |
| 9 | Exon | 18574 - 18723 | 13325690-13325839 | 150 | sequence | download |
| 9 | Intron | 18724 - 21302 | 13323111-13325689 | 2579 | sequence | - |
| 10 | Exon | 21303 - 21437 | 13322976-13323110 | 135 | sequence | download |
| 10 | Intron | 21438 - 24058 | 13320355-13322975 | 2621 | sequence | - |
| 11 | Exon | 24059 - 24617 | 13319796-13320354 | 559 | sequence | download |
| miRNAs |  |
|||
|---|---|---|---|---|
| Identifier | Name | Gene coordinates | Chromosomal coordinates | Prediction algorithm |
| Group 1 | ||||
| M1 | hsa-miR-653 | 24123 - 24142 | 13320271 - 13320290 | miranda |
| M2 | hsa-miR-384 | 24152 - 24174 | 13320239 - 13320261 | miranda |
| Group 2 | ||||
| M1 | hsa-miR-361-5p | 24456 - 24477 | 13319936 - 13319957 | miranda |
| M2 | hsa-miR-216b | 24491 - 24513 | 13319900 - 13319922 | miranda |















Please wait while loading ...