
| Gene |  |
||
|---|---|---|---|
| Symbol | SEC61A2 | ||
| Synonyms | FLJ10578 | ||
| Description | Sec61 alpha 2 subunit (S. cerevisiae) | ||
| EnsEMBL link | ENSG00000065665 | ||
| Chromosome | 10 - forward strand | ||
| Chromosomal location (UCSC link) |
12171640 - 12211956 (40317bp) | ||
| Sequence | sequence | ||
| Associated transcripts | NM_001142627 NM_001142628 NM_018144 AK301841 AK292295 BC026179 AF346603 AK057532 AK022640 AK022027 CR607709 AK001440 |
| Transcription start sites |  |
|||
|---|---|---|---|---|
| Identifier | Position | Gene coordinates | Chromosomal coordinates | Transcript |
| P1 | 1 | 1 | 12171640 | NM_001142627 NM_001142628 NM_018144 |
| P2 | 1 | 48 | 12171687 | AK301841 |
| P3 | 1 | 51 | 12171690 | AK292295 |
| P4 | 1 | 60 | 12171699 | AK057532 |
| P5 | 1 | 66 | 12171705 | AK022640 |
| P6 | 8 | 26146 | 12197785 | AK022027 |
| P7 | 9 | 27269 | 12198908 | AK001440 |
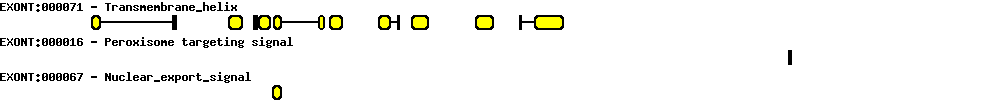
| Protein prediction |  |
|---|---|
| Transcript name | Sequence |
| NM_001142627 | sequence |
| NM_001142628 | sequence |
| NM_018144 | sequence |
| AK301841 | sequence |
| AK292295 | sequence |
| BC026179 | sequence |
| AF346603 | sequence |
| AK057532 | sequence |
| AK022640 | sequence |
| AK022027 | sequence |
| CR607709 | sequence |
| AK001440 | sequence |
| Polyadenylation |  |
||||||
|---|---|---|---|---|---|---|---|
| Identifier | Position | Gene coordinates of cleavage site | Chromosomal coordinates of cleavage site | Pattern | PolyA tail | Accession number | Type |
| PA1 | 13 | 34947 | 12206586 | no | AK301841 | Full_length | |
| PA2 | 13 | 35187 | 12206826 | AATAAA | no | AK022027 | Full_length |
| PA3 | 13 | 35187 | 12206826 | AATAAA | no | AK001440 | Full_length |
| PA4 | 13 | 35764 | 12207403 | AATAAA | no | NM_001142628 | Cdna |
| PA5 | 13 | 35764 | 12207403 | AATAAA | no | NM_018144 | Cdna |
| PA6 | 15 | 40317 | 12211956 | AATAAA | yes | NM_001142627 | Cdna |
| PA7 | 15 | 40317 | 12211956 | AATAAA | yes | BC026179 | Cdna |
| PA8 | 15 | 40317 | 12211956 | AATAAA | no | AK022640 | Full_length |
| PA9 | 15 | 41608 | 12213247 | ATTAAA | no | DB315342 | 3prime_est |
| Exon skipping |  |
|
|---|---|---|
| Identifier | Position | Transcript |
| ES1 | 3 | NM_001142628 AK301841 |
| ES2 | 4 | NM_001142627 NM_001142628 NM_018144 AK301841 AK292295 BC026179 AF346603 AK022640 |
| ES3 | 13 | NM_001142627 BC026179 AK057532 AK022640 |
| ES4 | 14 | NM_001142627 AK022640 |
| Conserved exons |  |
|
|---|---|---|
| Gene identifier mouse | Position human | Position mouse |
| 11395 | 1 | 1 |
| 11395 | 2 | 2 |
| 11395 | 3 | 3 |
| 11395 | 5 | 4 |
| 11395 | 6 | 5 |
| 11395 | 7 | 6 |
| 11395 | 8 | 7 |
| 11395 | 9 | 8 |
| 11395 | 10 | 9 |
| 11395 | 11 | 10 |
| 11395 | 12 | 11 |
| 11395 | 13 | 12 |
| Report |  |
|---|---|
| Exon | Number of supporting transcripts |
| Transcription initiation & first exon(s) | |
| 1 | 5 |
| 8 | 1 |
| 9 | 1 |
| Transcription termination & last exon(s) | |
| 13 | 5 |
| 15 | 4 |
| Exon skipping | |
| 3 | 2 |
| 4 | 8 |
| 13 | 4 |
| 14 | 2 |
| Expression |  |
||
|---|---|---|---|
| Exon position | Tissues | Cell lines | Cancer cell lines |
| 1 | Expression | Expression | Expression |
| 2 | Expression | Expression | Expression |
| 3 | Expression | Expression | Expression |
| 4 | Expression | Expression | Expression |
| 5 | Expression | Expression | Expression |
| 6 | Expression | Expression | Expression |
| 7 | Expression | Expression | Expression |
| 8 | Expression | Expression | Expression |
| 9 | Expression | Expression | Expression |
| 10 | Expression | Expression | Expression |
| 11 | Expression | Expression | Expression |
| 12 | Expression | Expression | Expression |
| 13 | Expression | Expression | Expression |
| 14 | Expression | Expression | Expression |
| 15 | Expression | Expression | Expression |
| Exon-Intron |  |
|||||
|---|---|---|---|---|---|---|
| Position | Type | Gene coordinates | Chromosomal coordinates | Length | Sequence | Features |
| 1 | Exon | 1 - 154 | 12171640 - 12171793 | 154 | sequence | download |
| 1 | Intron | 155 - 3600 | 12171794 - 12175239 | 3446 | sequence | - |
| 2 | Exon | 3601 - 3668 | 12175240 - 12175307 | 68 | sequence | download |
| 2 | Intron | 3669 - 6464 | 12175308 - 12178103 | 2796 | sequence | - |
| 3 | Exon | 6465 - 6530 | 12178104 - 12178169 | 66 | sequence | download |
| 3 | Intron | 6531 - 12792 | 12178170 - 12184431 | 6262 | sequence | - |
| 4 | Exon | 12793 - 12925 | 12184432 - 12184564 | 133 | sequence | download |
| 4 | Intron | 12926 - 13476 | 12184565 - 12185115 | 551 | sequence | - |
| 5 | Exon | 13477 - 13555 | 12185116 - 12185194 | 79 | sequence | download |
| 5 | Intron | 13556 - 19954 | 12185195 - 12191593 | 6399 | sequence | - |
| 6 | Exon | 19955 - 20086 | 12191594 - 12191725 | 132 | sequence | download |
| 6 | Intron | 20087 - 20211 | 12191726 - 12191850 | 125 | sequence | - |
| 7 | Exon | 20212 - 20321 | 12191851 - 12191960 | 110 | sequence | download |
| 7 | Intron | 20322 - 26137 | 12191961 - 12197776 | 5816 | sequence | - |
| 8 | Exon | 26138 - 26291 | 12197777 - 12197930 | 154 | sequence | download |
| 8 | Intron | 26292 - 27266 | 12197931 - 12198905 | 975 | sequence | - |
| 9 | Exon | 27267 - 27427 | 12198906 - 12199066 | 161 | sequence | download |
| 9 | Intron | 27428 - 28267 | 12199067 - 12199906 | 840 | sequence | - |
| 10 | Exon | 28268 - 28465 | 12199907 - 12200104 | 198 | sequence | download |
| 10 | Intron | 28466 - 31289 | 12200105 - 12202928 | 2824 | sequence | - |
| 11 | Exon | 31290 - 31481 | 12202929 - 12203120 | 192 | sequence | download |
| 11 | Intron | 31482 - 32572 | 12203121 - 12204211 | 1091 | sequence | - |
| 12 | Exon | 32573 - 32649 | 12204212 - 12204288 | 77 | sequence | download |
| 12 | Intron | 32650 - 34627 | 12204289 - 12206266 | 1978 | sequence | - |
| 13 | Exon | 34628 - 35764 | 12206267 - 12207403 | 1137 | sequence | download |
| 13 | Intron | 35765 - 38112 | 12207404 - 12209751 | 2348 | sequence | - |
| 14 | Exon | 38113 - 38253 | 12209752 - 12209892 | 141 | sequence | download |
| 14 | Intron | 38254 - 39617 | 12209893 - 12211256 | 1364 | sequence | - |
| 15 | Exon | 39618 - 40317 | 12211257 - 12211956 | 700 | sequence | download |

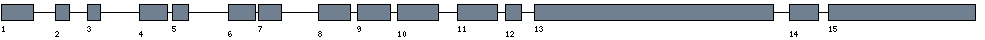

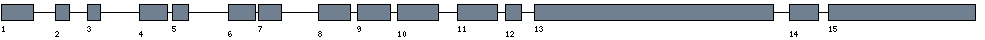

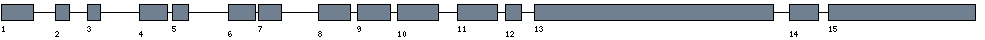

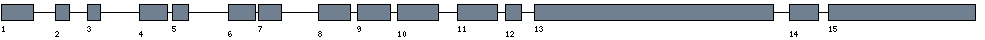

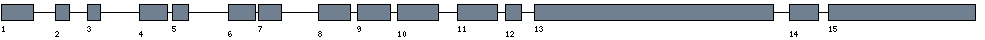
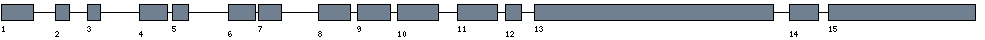

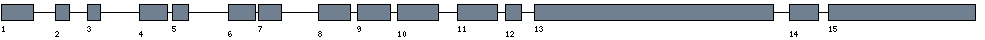
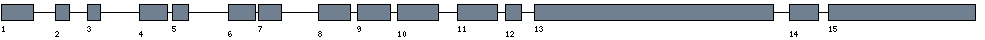

Please wait while loading ...